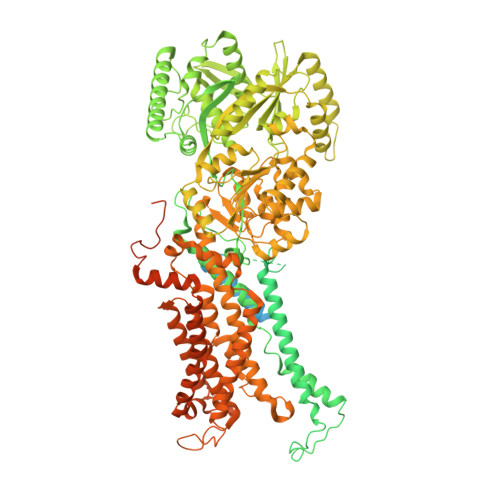
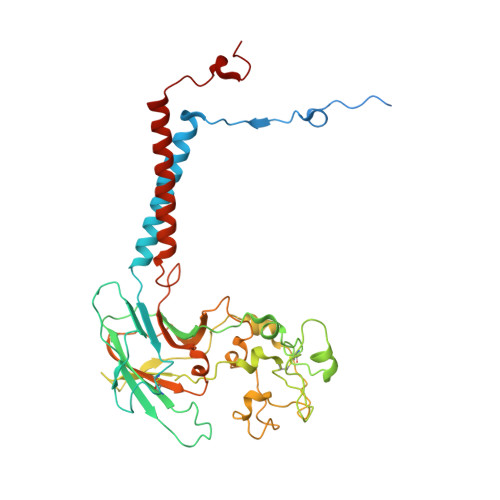
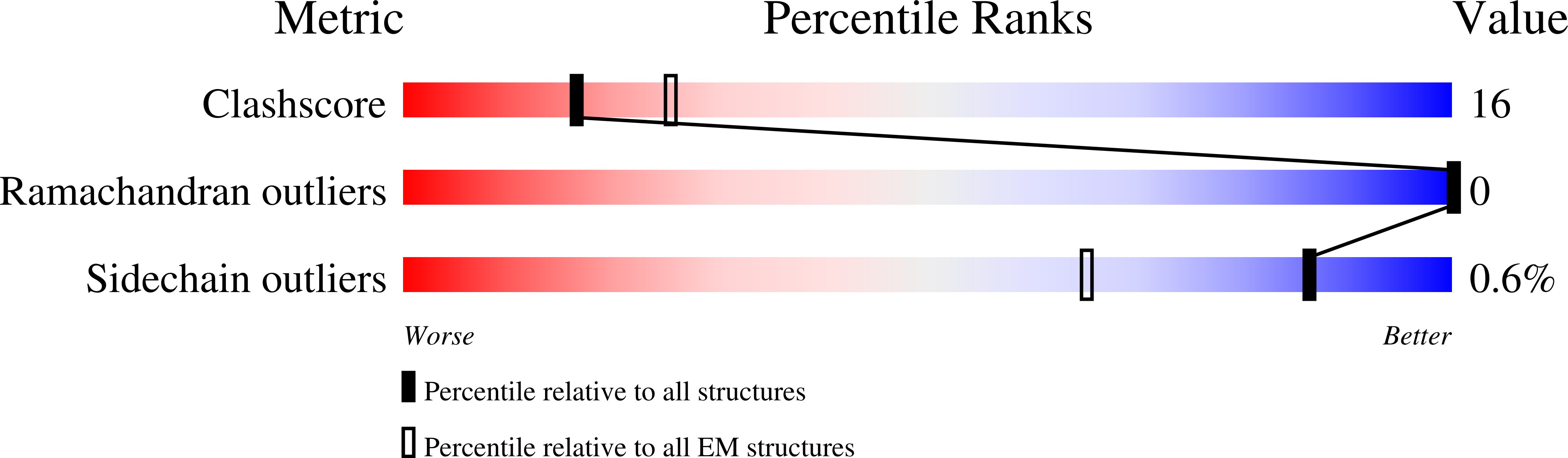Transport mechanism of P4 ATPase phosphatidylcholine flippases.
Bai, L., You, Q., Jain, B.K., Duan, H.D., Kovach, A., Graham, T.R., Li, H.(2020) Elife 9
- PubMed: 33320091
- DOI: https://doi.org/10.7554/eLife.62163
- Primary Citation of Related Structures:
7KY5, 7KY6, 7KY7, 7KY8, 7KY9, 7KYA, 7KYB, 7KYC - PubMed Abstract:
The P4 ATPases use ATP hydrolysis to transport large lipid substrates across lipid bilayers. The structures of the endosome- and Golgi-localized phosphatidylserine flippases-such as the yeast Drs2 and human ATP8A1-have recently been reported. However, a substrate-binding site on the cytosolic side has not been found, and the transport mechanisms of P4 ATPases with other substrates are unknown. Here, we report structures of the S. cerevisiae Dnf1-Lem3 and Dnf2-Lem3 complexes. We captured substrate phosphatidylcholine molecules on both the exoplasmic and cytosolic sides and found that they have similar structures. Unexpectedly, Lem3 contributes to substrate binding. The conformational transitions of these phosphatidylcholine transporters match those of the phosphatidylserine transporters, suggesting a conserved mechanism among P4 ATPases. Dnf1/Dnf2 have a unique P domain helix-turn-helix insertion that is important for function. Therefore, P4 ATPases may have retained an overall transport mechanism while evolving distinct features for different lipid substrates.
Organizational Affiliation:
Department of Biochemistry and Biophysics, School of Basic Medical Sciences, Peking University, Beijing, China.