Thiophene-Pyrazolourea Derivatives as Potent, Orally Bioavailable, and Isoform-Selective JNK3 Inhibitors.
Feng, Y., Park, H., Bauer, L., Ryu, J.C., Yoon, S.O.(2021) ACS Med Chem Lett 12: 24-29
- PubMed: 33488960
- DOI: https://doi.org/10.1021/acsmedchemlett.0c00533
- Primary Citation of Related Structures:
7KSI, 7KSJ, 7KSK - PubMed Abstract:
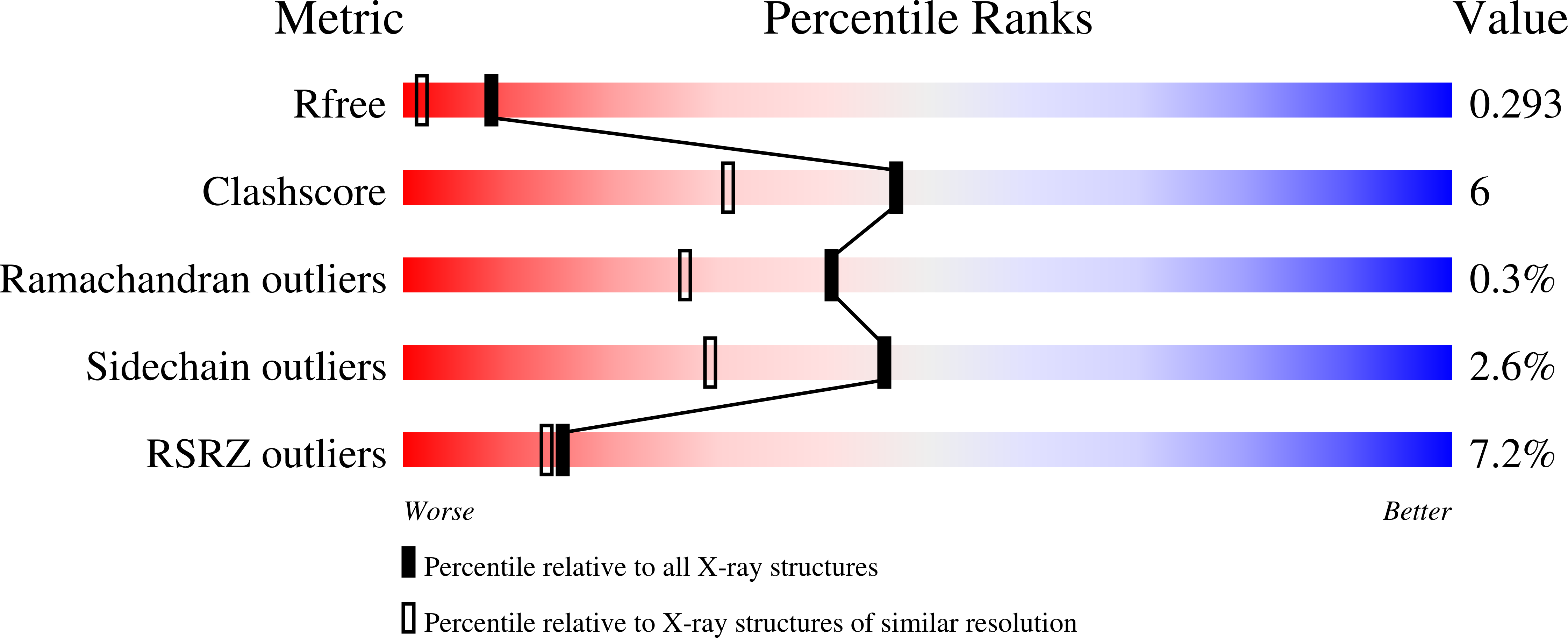
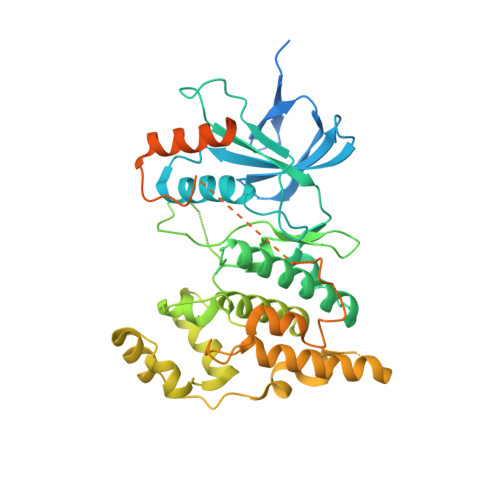
Potent JNK3 isoform selective inhibitors were developed from a thiophenyl-pyrazolourea scaffold. Through structure activity relationship (SAR) studies utilizing enzymatic and cell-based assays, and in vitro and in vivo drug metabolism and pharmacokinetic (DMPK) studies, potent and highly selective JNK3 inhibitors with oral bioavailability and brain penetrant capability were developed. Inhibitor 17 was a potent and isoform selective JNK3 inhibitor (IC 50 = 35 nM), had significant inhibition to only JNK3 in a panel profiling of 374 wild-type kinases, had high potency in functional cell-based assays, had high stability in human liver microsome ( t 1/2 = 66 min) and a clean CYP-450 inhibition profile, and was orally bioavailable and brain penetrant. Moreover, cocrystal structures of compounds 17 and 27 in human JNK3 were solved at 1.84 Å, which showed that these JNK3 isoform selective inhibitors bound to the ATP pocket, had interactions in both hydrophobic pocket-I and hydrophobic pocket-II.
Organizational Affiliation:
Reaction Biology Corporation, One Great Valley Parkway, Malvern, Pennsylvania 19355, United States.