An Aromatic Cluster in the Active Site of epi -Isozizaene Synthase Is an Electrostatic Toggle for Divergent Terpene Cyclization Pathways.
Ronnebaum, T.A., Gardner, S.M., Christianson, D.W.(2020) Biochemistry 59: 4744-4754
- PubMed: 33270439
- DOI: https://doi.org/10.1021/acs.biochem.0c00876
- Primary Citation of Related Structures:
7KJ8, 7KJ9, 7KJD, 7KJE, 7KJF, 7KJG - PubMed Abstract:
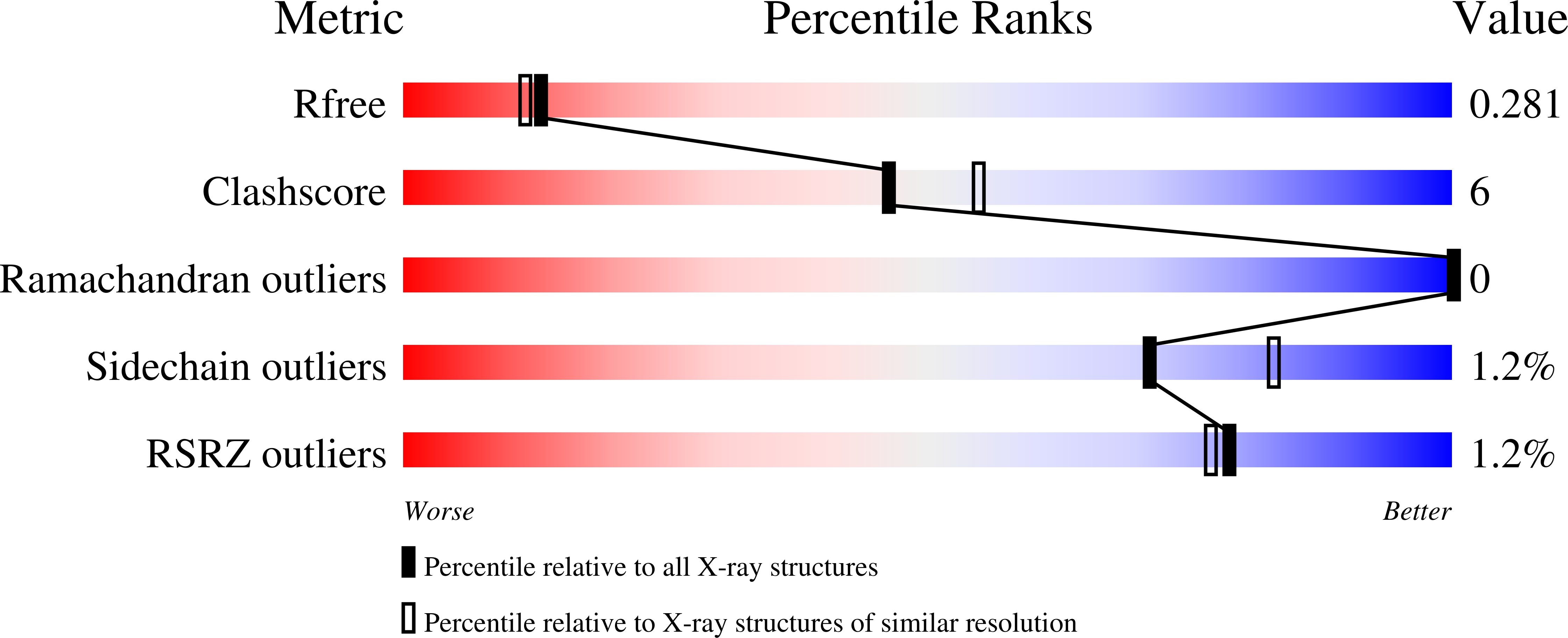
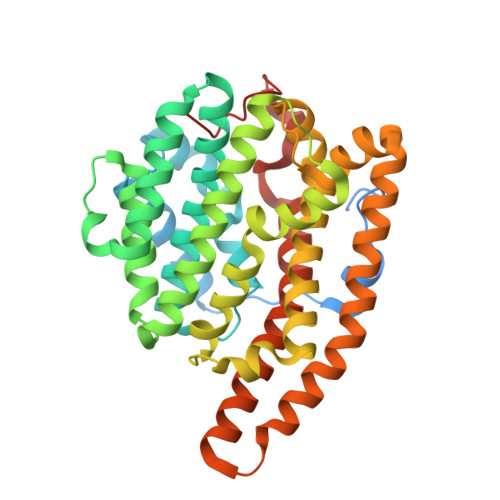
The sesquiterpene cyclase epi -isozizaene synthase (EIZS) catalyzes the cyclization of farnesyl diphosphate to form the tricyclic precursor of the antibiotic albaflavenone. The hydrophobic active site is largely defined by aromatic residues that direct a multistep reaction sequence through multiple carbocation intermediates. The previous substitution of polar residues for a key aromatic residue, F96, converts EIZS into a high-fidelity sesquisabinene synthase: the F96S, F96M, and F96Q variants generate 78%, 91%, and 97% sesquisabinene A, respectively. Here, we report high-resolution X-ray crystal structures of two of these reprogrammed cyclases. The structures of the F96M EIZS-Mg 2+ 3 -risedronate and F96M EIZS-Mg 2+ 3 -inorganic pyrophosphate-benzyltriethylammonium cation complexes reveal structural changes in the F96 aromatic cluster that redirect the cyclization pathway leading from the bisabolyl carbocation intermediate in catalysis. The structure of the F96S EIZS-Mg 2+ 3 -neridronate complex reveals a partially occupied inhibitor and an enzyme active site caught in transition between open and closed states. Finally, three structures of wild-type EIZS complexed with the bisphosphonate inhibitors neridronate, pamidronate, and risedronate provide a foundation for understanding binding differences between wild-type and variant enzymes. These structures provide new insight regarding active site flexibility, particularly with regard to the potential for subtle expansion and contraction to accommodate ligands of varying sizes as well as bound water molecules. Additionally, these structures highlight the importance of conformational changes in the F96 aromatic cluster that could influence cation-π interactions with carbocation intermediates in catalysis.
Organizational Affiliation:
Roy and Diana Vagelos Laboratories, Department of Chemistry, University of Pennsylvania, Philadelphia, Pennsylvania 19104-6323, United States.