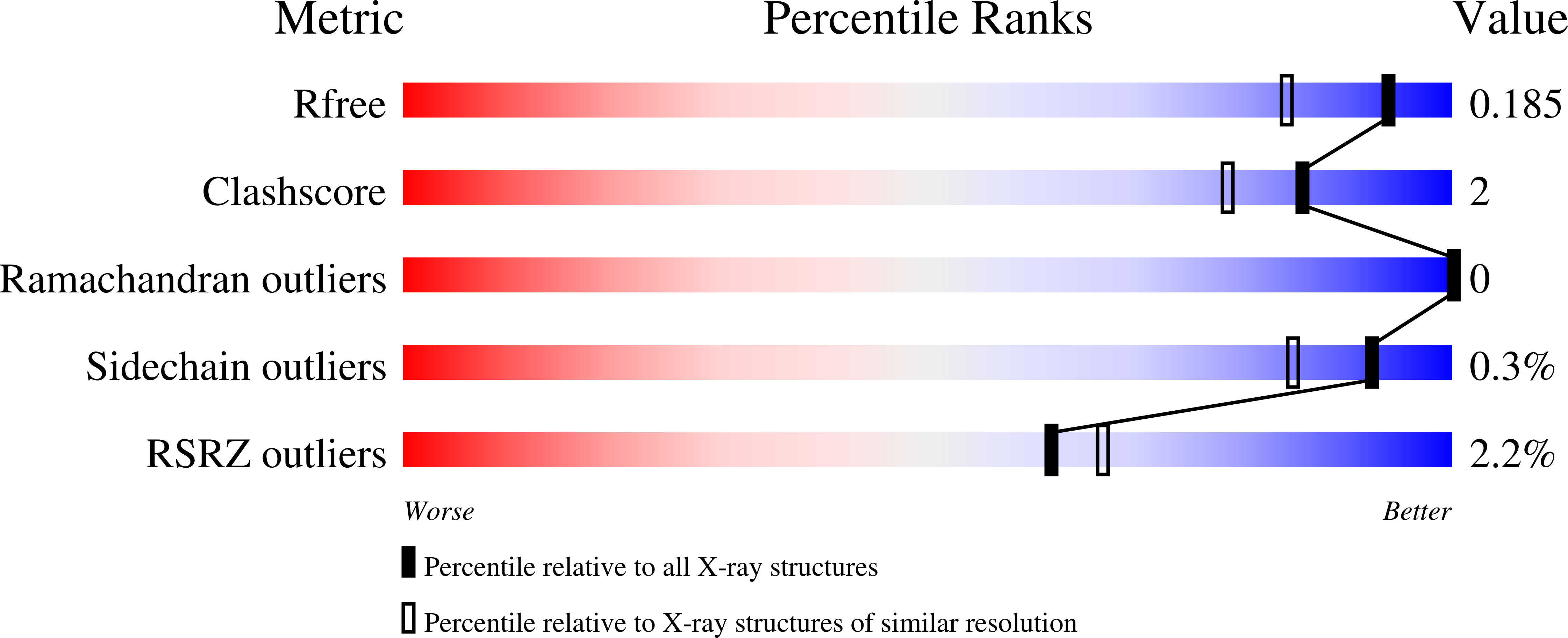
Structural analysis of histone deacetylase 8 mutants associated with Cornelia de Lange Syndrome spectrum disorders.
Osko, J.D., Porter, N.J., Decroos, C., Lee, M.S., Watson, P.R., Raible, S.E., Krantz, I.D., Deardorff, M.A., Christianson, D.W.(2020) J Struct Biol 213: 107681-107681
- PubMed: 33316326
- DOI: https://doi.org/10.1016/j.jsb.2020.107681
- Primary Citation of Related Structures:
7JVU, 7JVV, 7JVW - PubMed Abstract:
Cornelia de Lange Syndrome (CdLS) and associated spectrum disorders are characterized by one or more congenital anomalies including distinctive facial features, upper limb abnormalities, intellectual disability, and other symptoms. The molecular genetic basis of CdLS is linked to defects in cohesin, a protein complex that functions in sister chromatid cohesion, chromatin organization, and transcriptional regulation. Histone deacetylase 8 (HDAC8) plays an important role in cohesin function by catalyzing the deacetylation of SMC3, which is required for efficient recycling of the cohesin complex. Missense mutations in HDAC8 have been identified in children diagnosed with CdLS spectrum disorders, and here we outline structure-function relationships for four of these mutations. Specifically, we report the 1.50 Å-resolution structure of the I45T HDAC8-suberoylanilide hydroxamic acid complex, the 1.84 Å-resolution structure of E66D/Y306F HDAC8 complexed with a peptide assay substrate, and the 2.40 Å-resolution structure of G320R HDAC8 complexed with the inhibitor M344. Additionally, we present a computationally generated model of D176G HDAC8. These structures illuminate new structure-function relationships for HDAC8 and highlight the importance of long-range interactions in the protein scaffold that can influence catalytic function.
Organizational Affiliation:
Roy and Diana Vagelos Laboratories, Department of Chemistry, University of Pennsylvania, 231 South 34(th) Street, Philadelphia, PA 19104-6323, United States.