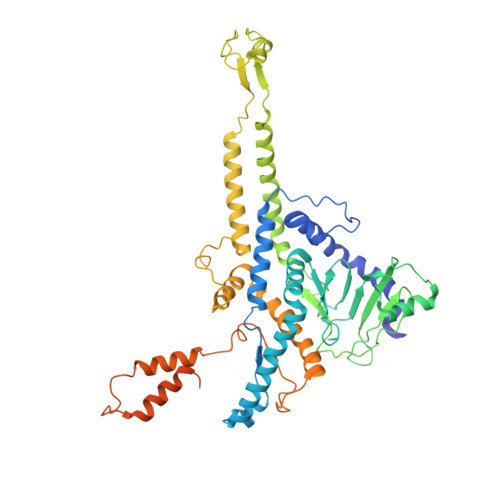
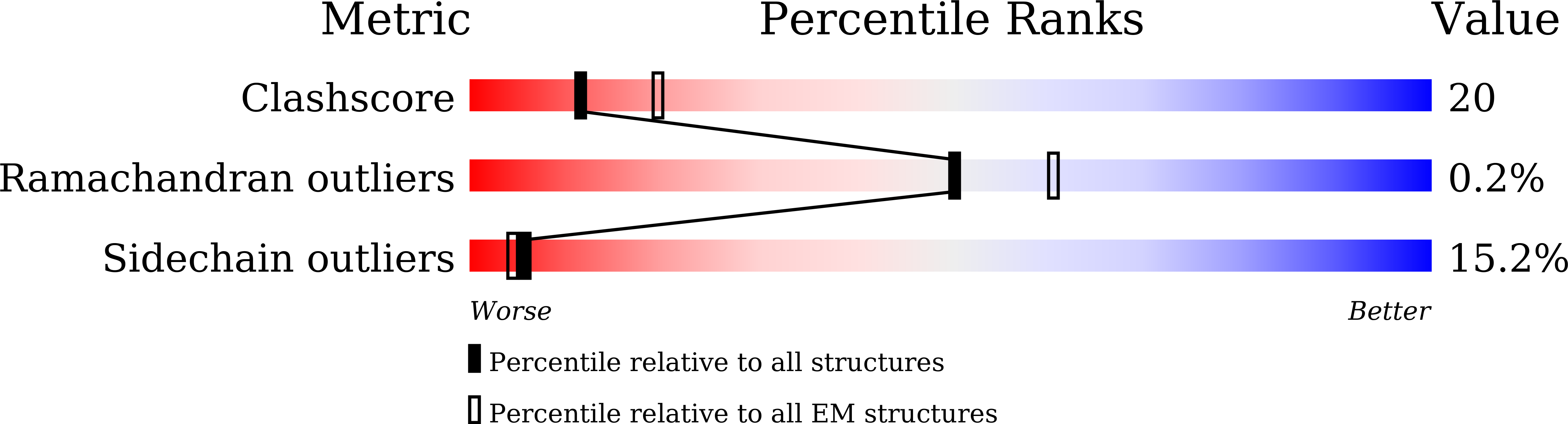Structural changes in bacteriophage T7 upon receptor-induced genome ejection.
Chen, W., Xiao, H., Wang, L., Wang, X., Tan, Z., Han, Z., Li, X., Yang, F., Liu, Z., Song, J., Liu, H., Cheng, L.(2021) Proc Natl Acad Sci U S A 118
- PubMed: 34504014
- DOI: https://doi.org/10.1073/pnas.2102003118
- Primary Citation of Related Structures:
7EY6, 7EY7, 7EY8, 7EY9, 7EYB - PubMed Abstract:
Many tailed bacteriophages assemble ejection proteins and a portal-tail complex at a unique vertex of the capsid. The ejection proteins form a transenvelope channel extending the portal-tail channel for the delivery of genomic DNA in cell infection. Here, we report the structure of the mature bacteriophage T7, including the ejection proteins, as well as the structures of the full and empty T7 particles in complex with their cell receptor lipopolysaccharide. Our near-atomic-resolution reconstruction shows that the ejection proteins in the mature T7 assemble into a core, which comprises a fourfold gene product 16 (gp16) ring, an eightfold gp15 ring, and a putative eightfold gp14 ring. The gp15 and gp16 are mainly composed of helix bundles, and gp16 harbors a lytic transglycosylase domain for degrading the bacterial peptidoglycan layer. When interacting with the lipopolysaccharide, the T7 tail nozzle opens. Six copies of gp14 anchor to the tail nozzle, extending the nozzle across the lipopolysaccharide lipid bilayer. The structures of gp15 and gp16 in the mature T7 suggest that they should undergo remarkable conformational changes to form the transenvelope channel. Hydrophobic α-helices were observed in gp16 but not in gp15, suggesting that gp15 forms the channel in the hydrophilic periplasm and gp16 forms the channel in the cytoplasmic membrane.
Organizational Affiliation:
Key Laboratory for Matter Microstructure and Function of Hunan Province, Key Laboratory of Low-dimensional Quantum Structures and Quantum Control, School of Physics and Electronics, Hunan Normal University, Changsha 410082, China.