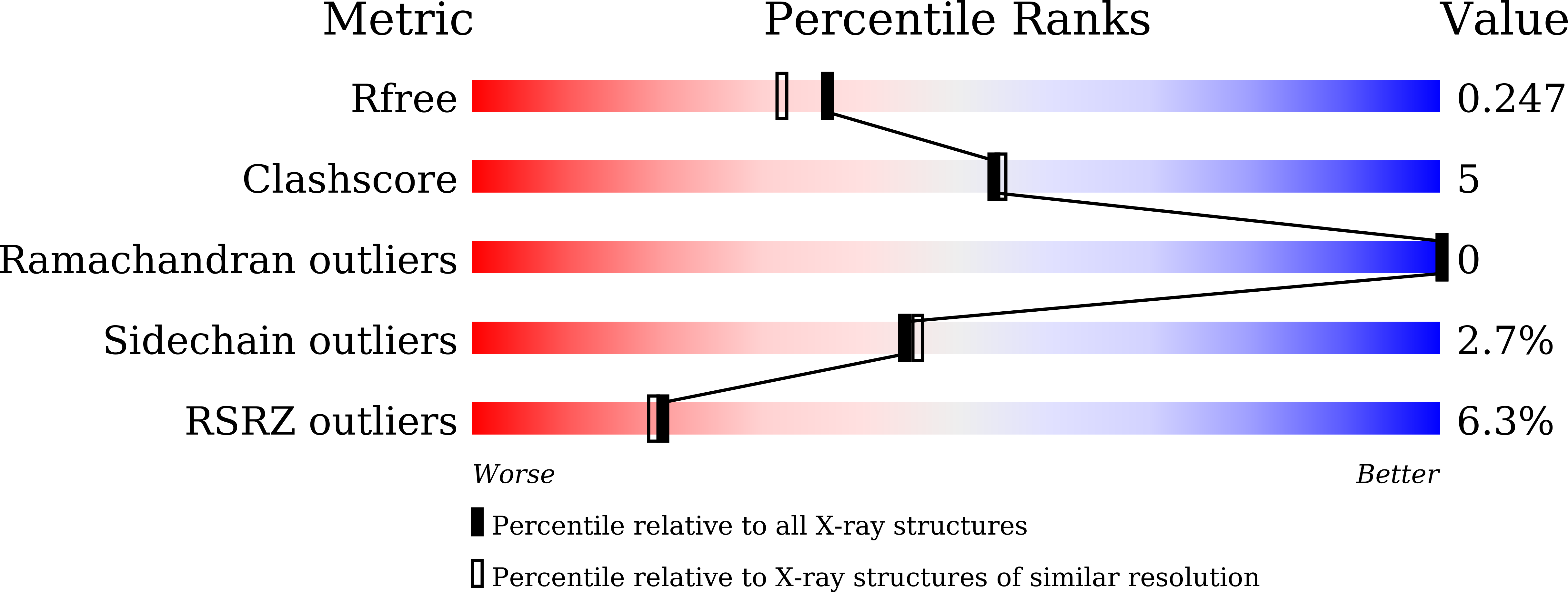
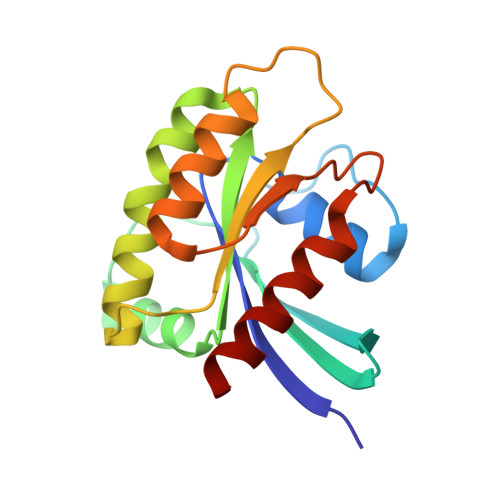
KRAS(G12D) can be targeted by potent inhibitors via formation of salt bridge.
Mao, Z., Xiao, H., Shen, P., Yang, Y., Xue, J., Yang, Y., Shang, Y., Zhang, L., Li, X., Zhang, Y., Du, Y., Chen, C.C., Guo, R.T., Zhang, Y.(2022) Cell Discov 8: 5-5
- PubMed: 35075146
- DOI: https://doi.org/10.1038/s41421-021-00368-w
- Primary Citation of Related Structures:
7EW9, 7EWA, 7EWB - PubMed Abstract:
KRAS mutation occurs in nearly 30% of human cancers, yet the most prevalent and oncogenic KRAS(G12D) variant still lacks inhibitors. Herein, we designed a series of potent inhibitors that can form a salt bridge with KRAS's Asp12 residue. Our ITC results show that these inhibitors have similar binding affinity with both GDP-bound and GTP-bound KRAS(G12D), and our crystallographic studies reveal the structural basis of inhibitor binding-induced switch-II pocket in KRAS(G12D), experimentally confirming the formation of a salt bridge between the piperazine moiety of the inhibitors and the Asp12 residue of the mutant protein. Among KRAS family proteins and mutants, both ITC and enzymatic assays demonstrate the selectivity of the inhibitors for KRAS(G12D); and the inhibitors disrupt the KRAS-CRAF interaction. We also observed the inhibition of cancer cell proliferation as well as MAPK signaling by a representative inhibitor (TH-Z835). However, since the inhibition was not fully dependent on KRAS mutation status, it is possible that our inhibitors may have off-target effects via targeting non-KRAS small GTPases. Experiments with mouse xenograft models of pancreatic cancer showed that TH-Z835 significantly reduced tumor volume and synergized with an anti-PD-1 antibody. Collectively, our study demonstrates proof-of-concept for a strategy based on salt-bridge and induced-fit pocket formation for KRAS(G12D) targeting, which warrants future medicinal chemistry efforts for optimal efficacy and minimized off-target effects.
Organizational Affiliation:
School of Pharmaceutical Science, Tsinghua-Peking Center for Life Sciences, Tsinghua University; Beijing Advanced Innovation Center for Human Brain Protection, Beijing, China.