Inhibition of Mycobacterium tuberculosis InhA by 3-nitropropanoic acid.
Songsiriritthigul, C., Hanwarinroj, C., Pakamwong, B., Srimanote, P., Suttipanta, N., Sureram, S., Suttisintong, K., Kamsri, P., Punkvang, A., Spencer, J., Kittakoop, P., Pungpo, P.(2022) Proteins 90: 898-904
- PubMed: 34677871
- DOI: https://doi.org/10.1002/prot.26268
- Primary Citation of Related Structures:
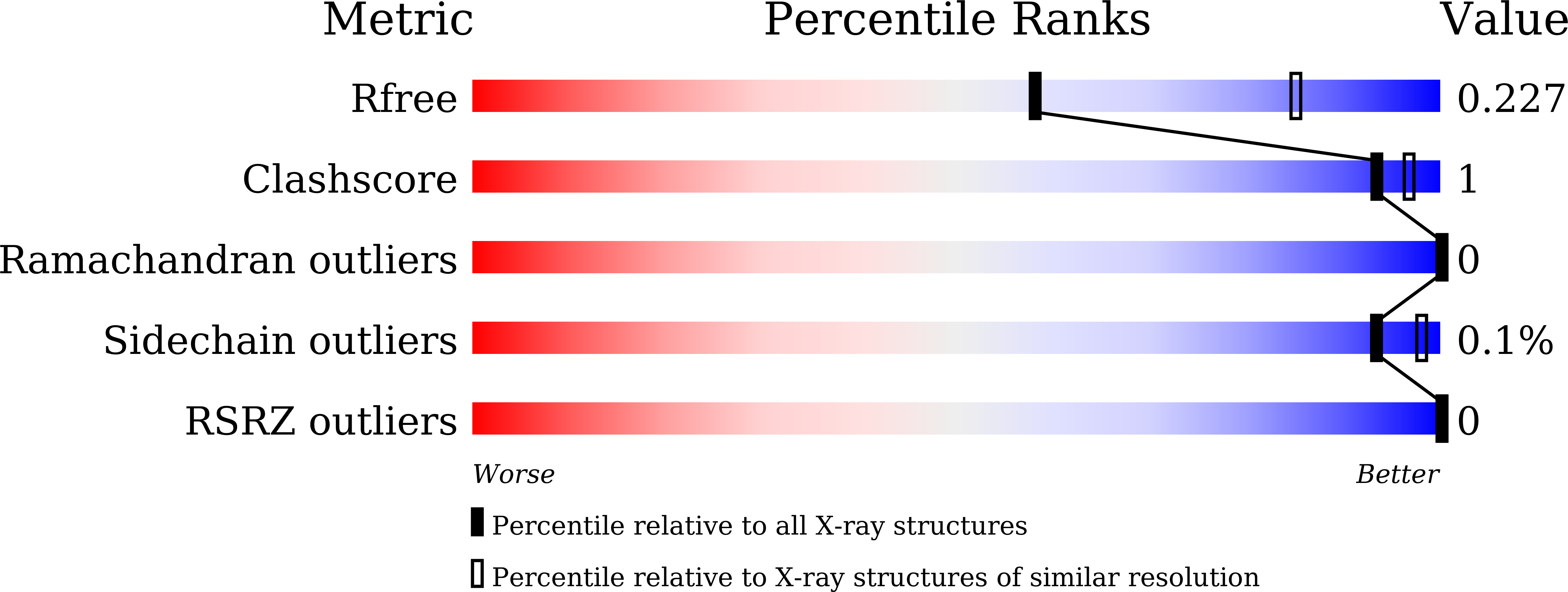
7E48 - PubMed Abstract:
3-Nitropropanoic acid (3NP), a bioactive fungal natural product, was previously demonstrated to inhibit growth of Mycobacterium tuberculosis. Here we demonstrate that 3NP inhibits the 2-trans-enoyl-acyl carrier protein reductase (InhA) from Mycobacterium tuberculosis with an IC 50 value of 71 μM, and present the crystal structure of the ternary InhA-NAD + -3NP complex. The complex contains the InhA substrate-binding loop in an ordered, open conformation with Tyr158, a catalytically important residue whose orientation defines different InhA substrate/inhibitor complex conformations, in the "out" position. 3NP occupies a hydrophobic binding site adjacent to the NAD + cofactor and close to that utilized by the diphenyl ether triclosan, but binds predominantly via electrostatic and water-mediated hydrogen-bonding interactions with the protein backbone and NAD + cofactor. The identified mode of 3NP binding provides opportunities to improve inhibitory activity toward InhA.
Organizational Affiliation:
Synchrotron Light Research Institute (Public Organization), Nakhon Ratchasima, Thailand.