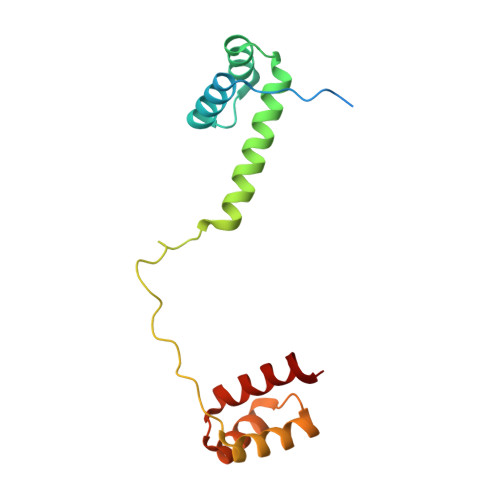
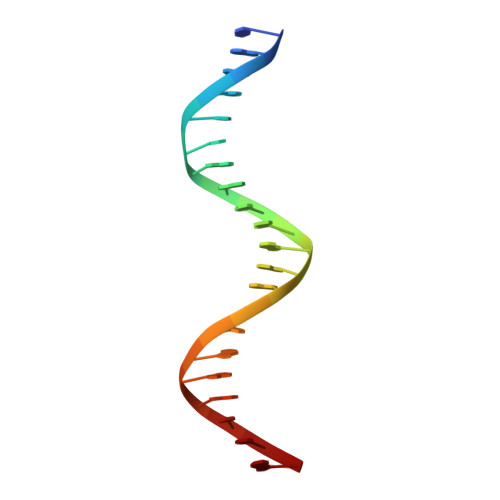
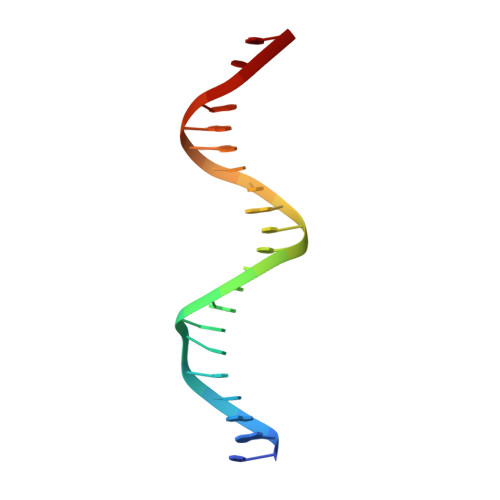
DNA crosslinking and recombination-activating genes 1/2 (RAG1/2) are required for oncogenic splicing in acute lymphoblastic leukemia.
Zhang, H., Cheng, N., Li, Z., Bai, L., Fang, C., Li, Y., Zhang, W., Dong, X., Jiang, M., Liang, Y., Zhang, S., Mi, J., Zhu, J., Zhang, Y., Chen, S.J., Zhao, Y., Weng, X.Q., Hu, W., Chen, Z., Huang, J., Meng, G.(2021) Cancer Commun (Lond) 41: 1116-1136
- PubMed: 34699692
- DOI: https://doi.org/10.1002/cac2.12234
- Primary Citation of Related Structures:
7DW5 - PubMed Abstract:
Abnormal alternative splicing is frequently associated with carcinogenesis. In B-cell acute lymphoblastic leukemia (B-ALL), double homeobox 4 fused with immunoglobulin heavy chain (DUX4/IGH) can lead to the aberrant production of E-26 transformation-specific family related gene abnormal transcript (ERG alt ) and other splicing variants. However, the molecular mechanism underpinning this process remains elusive. Here, we aimed to know how DUX4/IGH triggers abnormal splicing in leukemia. The differential intron retention analysis was conducted to identify novel DUX4/IGH-driven splicing in B-ALL patients. X-ray crystallography, small angle X-ray scattering (SAXS), and analytical ultracentrifugation were used to investigate how DUX4/IGH recognize double DUX4 responsive element (DRE)-DRE sites. The ERG alt biogenesis and B-cell differentiation assays were performed to characterize the DUX4/IGH crosslinking activity. To check whether recombination-activating gene 1/2 (RAG1/2) was required for DUX4/IGH-driven splicing, the proximity ligation assay, co-immunoprecipitation, mammalian two hybrid characterizations, in vitro RAG1/2 cleavage, and shRNA knock-down assays were performed. We reported previously unrecognized intron retention events in C-type lectin domain family 12, member A abnormal transcript (CLEC12A alt ) and chromosome 6 open reading frame 89 abnormal transcript (C6orf89 alt ), where also harbored repetitive DRE-DRE sites. Supportively, X-ray crystallography and SAXS characterization revealed that DUX4 homeobox domain (HD)1-HD2 might dimerize into a dumbbell-shape trans configuration to crosslink two adjacent DRE sites. Impaired DUX4/IGH-mediated crosslinking abolishes ERG alt , CLEC12A alt , and C6orf89 alt biogenesis, resulting in marked alleviation of its inhibitory effect on B-cell differentiation. Furthermore, we also observed a rare RAG1/2-mediated recombination signal sequence-like DNA edition in DUX4/IGH target genes. Supportively, shRNA knock-down of RAG1/2 in leukemic Reh cells consistently impaired the biogenesis of ERG alt , CLEC12A alt , and C6orf89 alt . All these results suggest that DUX4/IGH-driven DNA crosslinking is required for RAG1/2 recruitment onto the double tandem DRE-DRE sites, catalyzing V(D)J-like recombination and oncogenic splicing in acute lymphoblastic leukemia.
Organizational Affiliation:
Shanghai Institute of Hematology, State Key Laboratory of Medical Genomics, National Research Center for Translational Medicine, Rui-Jin Hospital, School of Medicine and School of Life Sciences and Biotechnology, Shanghai JiaoTong University, Shanghai, 200025, P. R. China.