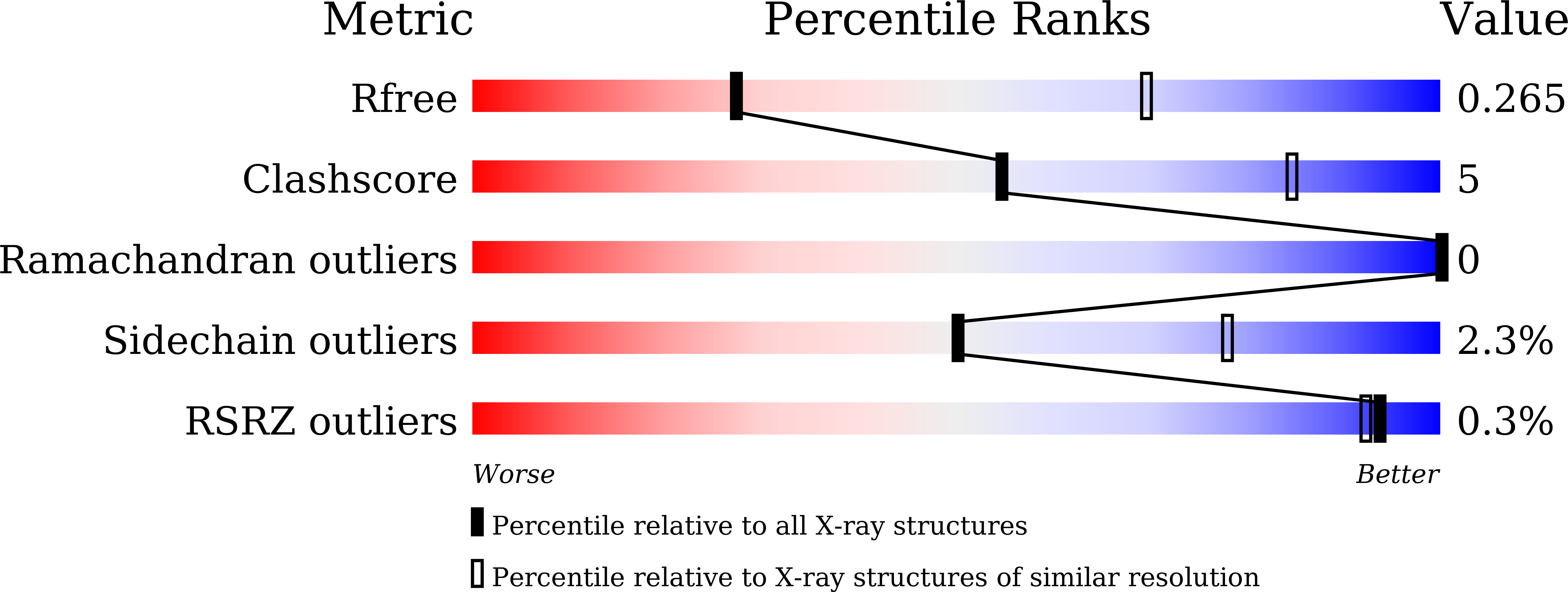
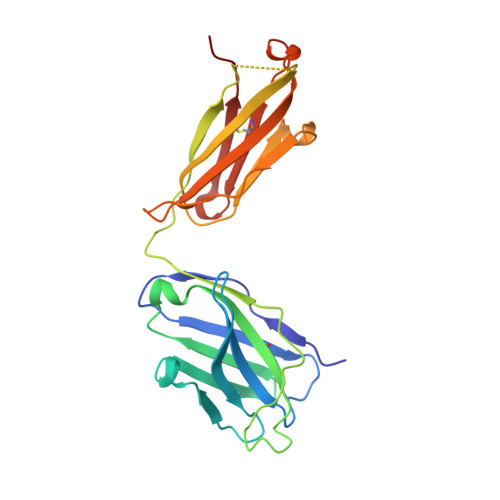
Antibody-dependent enhancement (ADE) of SARS-CoV-2 pseudoviral infection requires Fc gamma RIIB and virus-antibody complex with bivalent interaction.
Wang, S., Wang, J., Yu, X., Jiang, W., Chen, S., Wang, R., Wang, M., Jiao, S., Yang, Y., Wang, W., Chen, H., Chen, B., Gu, C., Liu, C., Wang, A., Wang, M., Li, G., Guo, C., Liu, D., Zhang, J., Zhang, M., Wang, L., Gui, X.(2022) Commun Biol 5: 262-262
- PubMed: 35332252
- DOI: https://doi.org/10.1038/s42003-022-03207-0
- Primary Citation of Related Structures:
7DJZ, 7DK0 - PubMed Abstract:
Understanding the underlying molecular mechanisms behind ADE of SARS-CoV-2 is critical for development of safe and effective therapies. Here, we report that two neutralizing mAbs, MW01 and MW05, could enhance the infection of SARS-CoV-2 pseudovirus on FcγRIIB-expressing B cells. X-ray crystal structure determination and S trimer-binding modeling showed that MW01 and MW05 could bind to RBDs in S trimer with both "up" and "down" states. While, the neutralizing mAb MW07, which has no ADE activity only binds to RBD in S trimer with "up" state. Monovalent MW01 and MW05 completely diminished the ADE activity compared with their bivalent counterparts. Moreover, both macropinocytosis and endocytosis are confirmed involving in ADE of SARS-CoV-2 pseudoviral infection. Blocking endosome transportation and lysosome acidification could inhibit the ADE activity mediated by MW05. Together, our results identified a novel ADE mechanism of SARS-CoV-2 pseudovirus in vitro, FcγRIIB-mediated uptake of SARS-CoV-2/mAb complex with bivalent interaction.
Organizational Affiliation:
Mabwell (Shanghai) Bioscience Co., Ltd, Shanghai, 201210, China.