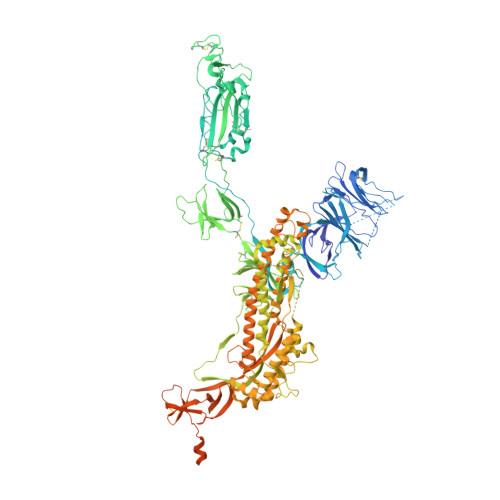
Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Yan, R., Wang, R., Ju, B., Yu, J., Zhang, Y., Liu, N., Wang, J., Zhang, Q., Chen, P., Zhou, B., Li, Y., Shen, Y., Zhang, S., Tian, L., Guo, Y., Xia, L., Zhong, X., Cheng, L., Ge, X., Zhao, J., Wang, H.W., Wang, X., Zhang, Z., Zhang, L., Zhou, Q.(2021) Cell Res 31: 517-525
- PubMed: 33731853
- DOI: https://doi.org/10.1038/s41422-021-00487-9
- Primary Citation of Related Structures:
7CZP, 7CZQ, 7CZR, 7CZS, 7CZT, 7CZU, 7CZV, 7CZW, 7CZX, 7CZY, 7CZZ, 7D00, 7D03, 7D0B, 7D0C, 7D0D - PubMed Abstract:
Neutralizing monoclonal antibodies (nAbs) to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) represent promising candidates for clinical intervention against coronavirus disease 2019 (COVID-19). We isolated a large number of nAbs from SARS-CoV-2-infected individuals capable of disrupting proper interaction between the receptor binding domain (RBD) of the viral spike (S) protein and the receptor angiotensin converting enzyme 2 (ACE2). However, the structural basis for their potent neutralizing activity remains unclear. Here, we report cryo-EM structures of the ten most potent nAbs in their native full-length IgG-form or in both IgG-form and Fab-form bound to the trimeric S protein of SARS-CoV-2. The bivalent binding of the full-length IgG is found to associate with more RBDs in the "up" conformation than the monovalent binding of Fab, perhaps contributing to the enhanced neutralizing activity of IgG and triggering more shedding of the S1 subunit from the S protein. Comparison of a large number of nAbs identified common and unique structural features associated with their potent neutralizing activities. This work provides a structural basis for further understanding the mechanism of nAbs, especially through revealing the bivalent binding and its correlation with more potent neutralization and the shedding of S1 subunit.
Organizational Affiliation:
Center for Infectious Disease Research, Westlake Laboratory of Life Sciences and Biomedicine, Key Laboratory of Structural Biology of Zhejiang Province, School of Life Sciences, Westlake University, 18 Shilongshan Road, Hangzhou, Zhejiang, 310024, China.