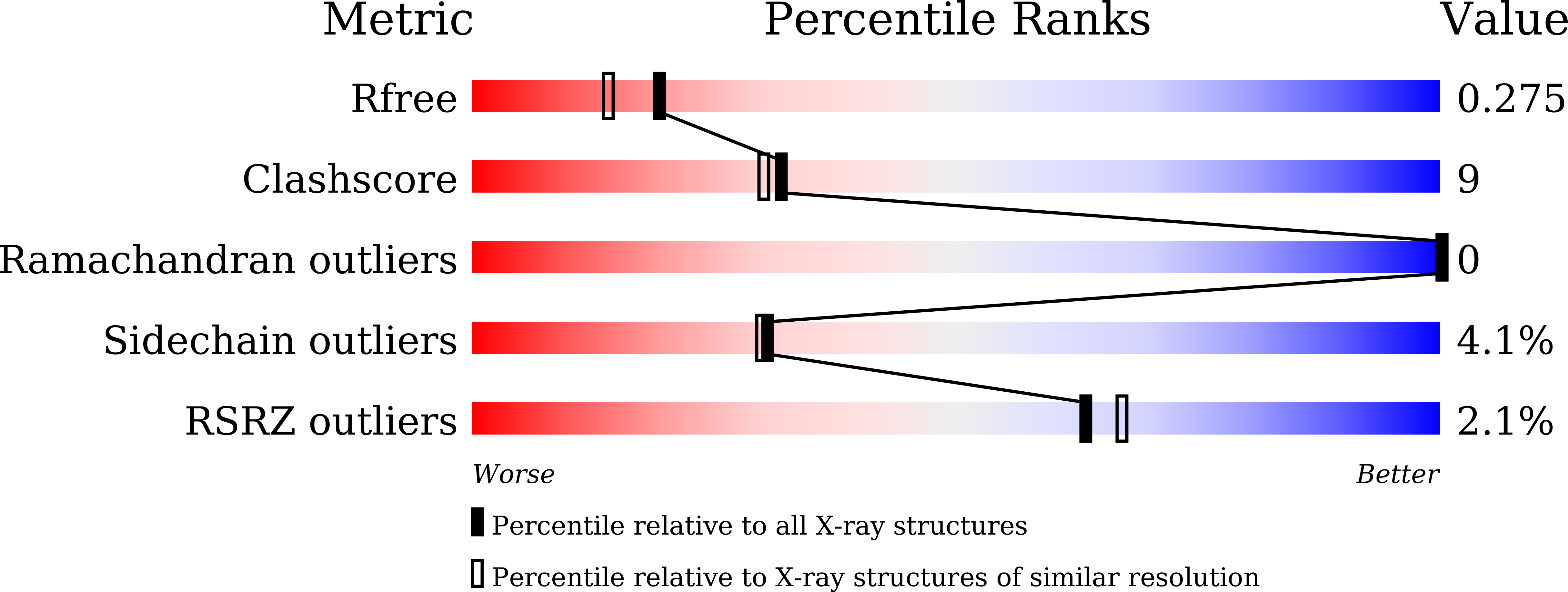
Crystal structure of a novel homodimeric l-ribulose 3-epimerase from Methylomonus sp.
Yoshida, H., Yoshihara, A., Kato, S., Mochizuki, S., Akimitsu, K., Izumori, K., Kamitori, S.(2021) FEBS Open Bio 11: 1621-1637
- PubMed: 33838083
- DOI: https://doi.org/10.1002/2211-5463.13159
- Primary Citation of Related Structures:
7CJ4, 7CJ5, 7CJ6, 7CJ7, 7CJ8, 7CJ9 - PubMed Abstract:
d-Allulose has potential as a low-calorie sweetener which can suppress fat accumulation. Several enzymes capable of d-allulose production have been isolated, including d-tagatose 3-epimerases. Here, we report the isolation of a novel protein from Methylomonas sp. expected to be a putative enzyme based on sequence similarity to ketose 3-epimerase. The synthesized gene encoding the deduced ketose 3-epimerase was expressed as a recombinant enzyme in Escherichia coli, and it exhibited the highest enzymatic activity toward l-ribulose, followed by d-ribulose and d-allulose. The X-ray structure analysis of l-ribulose 3-epimerase from Methylomonas sp. (MetLRE) revealed a homodimeric enzyme, the first reported structure of dimeric l-ribulose 3-epimerase. The monomeric structure of MetLRE is similar to that of homotetrameric l-ribulose 3-epimerases, but the short C-terminal α-helix of MetLRE is unique and different from those of known l-ribulose 3 epimerases. The length of the C-terminal α-helix was thought to be involved in tetramerization and increasing stability; however, the addition of residues to MetLRE at the C terminus did not lead to tetramer formation. MetLRE is the first dimeric l-ribulose 3-epimerase identified to exhibit high relative activity toward d-allulose.
Organizational Affiliation:
Life Science Research Center and Faculty of Medicine, Kagawa University, Kita, Japan.