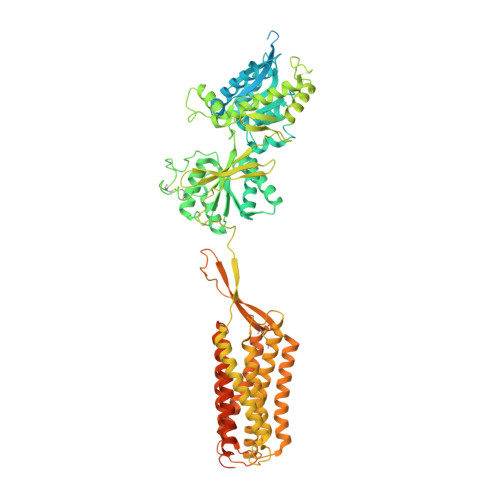
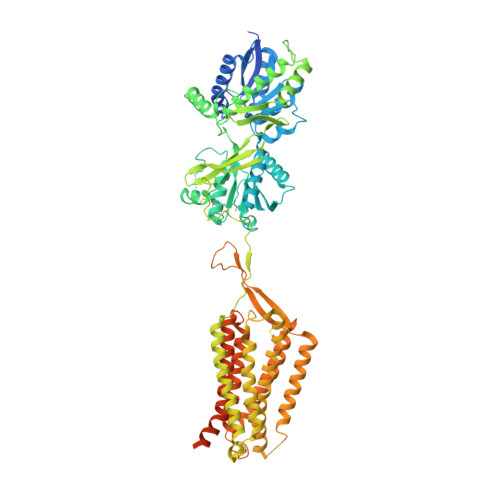
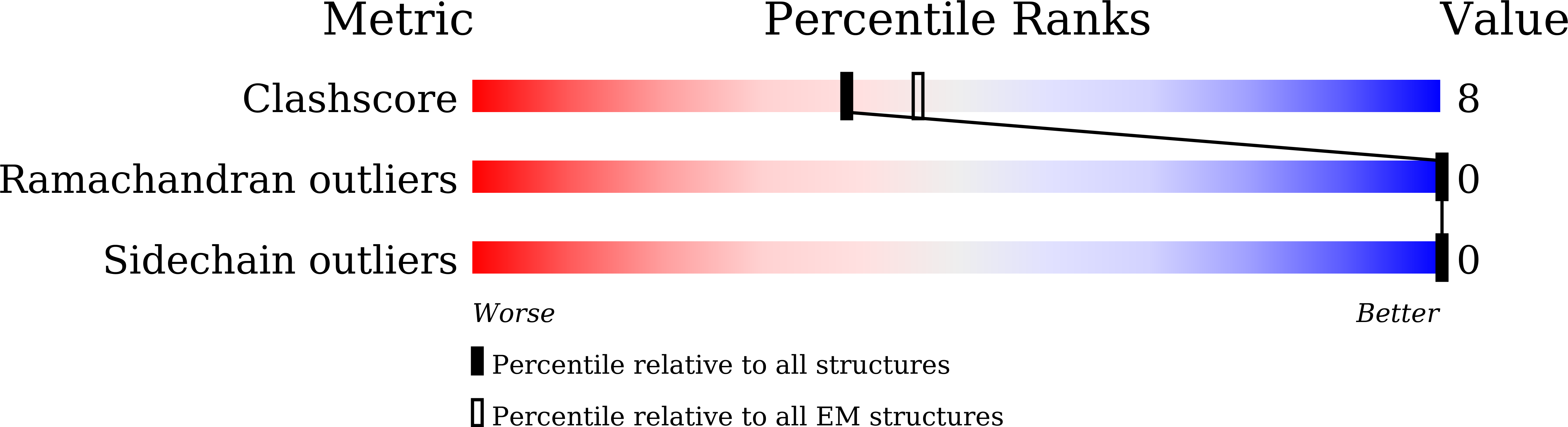Cryo-EM structures of inactive and active GABABreceptor.
Mao, C., Shen, C., Li, C., Shen, D.D., Xu, C., Zhang, S., Zhou, R., Shen, Q., Chen, L.N., Jiang, Z., Liu, J., Zhang, Y.(2020) Cell Res 30: 564-573
- PubMed: 32494023
- DOI: https://doi.org/10.1038/s41422-020-0350-5
- Primary Citation of Related Structures:
7C7Q, 7C7S - PubMed Abstract:
Metabotropic GABA B G protein-coupled receptor functions as a mandatory heterodimer of GB1 and GB2 subunits and mediates inhibitory neurotransmission in the central nervous system. Each subunit is composed of the extracellular Venus flytrap (VFT) domain and transmembrane (TM) domain. Here we present cryo-EM structures of full-length human heterodimeric GABA B receptor in the antagonist-bound inactive state and in the active state complexed with an agonist and a positive allosteric modulator in the presence of G i1 protein at a resolution range of 2.8-3.0 Å. Our structures reveal that agonist binding stabilizes the closure of GB1 VFT, which in turn triggers a rearrangement of TM interfaces between the two subunits from TM3-TM5/TM3-TM5 in the inactive state to TM6/TM6 in the active state and finally induces the opening of intracellular loop 3 and synergistic shifting of TM3, 4 and 5 helices in GB2 TM domain to accommodate the α5-helix of G i1 . We also observed that the positive allosteric modulator anchors at the dimeric interface of TM domains. These results provide a structural framework for understanding class C GPCR activation and a rational template for allosteric modulator design targeting the dimeric interface of GABA B receptor.
Organizational Affiliation:
Department of Biophysics, and Department of Pathology of Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, 310058, Zhejiang, China.