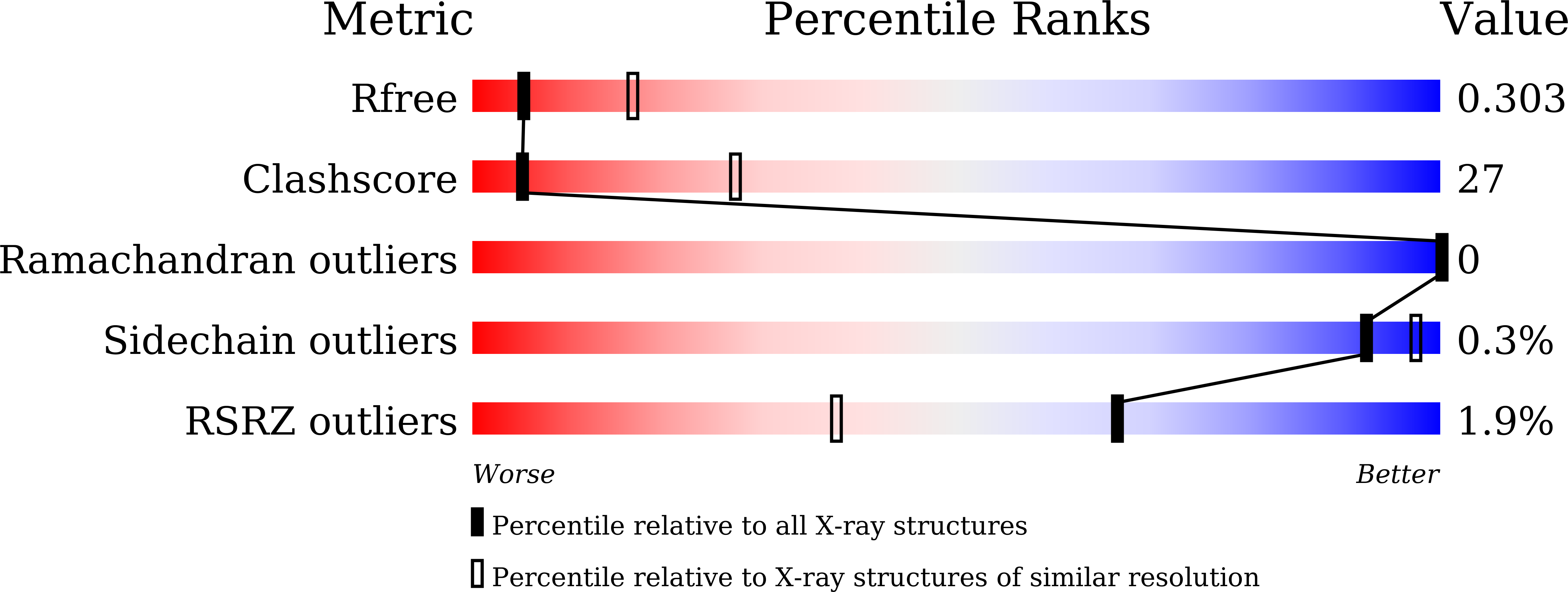
Design and Synthesis of TRAP1 Selective Inhibitors: H-Bonding with Asn171 Residue in TRAP1 Increases Paralog Selectivity.
Yang, S., Yoon, N.G., Kim, D., Park, E., Kim, S.Y., Lee, J.H., Lee, C., Kang, B.H., Kang, S.(2021) ACS Med Chem Lett 12: 1173-1180
- PubMed: 34267888
- DOI: https://doi.org/10.1021/acsmedchemlett.1c00213
- Primary Citation of Related Structures:
7C7B, 7C7C - PubMed Abstract:
Tumor necrosis factor receptor-associated protein 1 (TRAP1) is overexpressed in the mitochondria of various cancer cells, reprograms cellular metabolism to enable cancer cells to adapt to harsh tumor environments. As inactivation of TRAP1 induces massive apoptosis in cancer cells in vitro and in vivo , the development of TRAP1-selective inhibitors has become an attractive approach. A series of purine-8-one and pyrrolo[2,3- d ]pyrimidine derivatives was developed based on TRAP1 structure and identified to be highly selective in vitro for TRAP1 over the paralogous enzymes, Hsp90α and Grp94. The TRAP1-selective inhibition strategy via utilization of the Asn171 residue of the ATP-lid was investigated using X-ray crystallography and molecular dynamics simulation studies. Among various synthesized potent TRAP1 inhibitors, 5f possessed a 65-fold selectivity over Hsp90α and a 13-fold selectivity over Grp94. Additionally, 6f had a half-maximal inhibitory concentration (IC 50 ) of 63.5 nM for TRAP1, with a 78-fold and 30-fold selectivity over Hsp90α and Grp94, respectively.
Organizational Affiliation:
College of Pharmacy, Ewha Womans University, Seoul 03760, Republic of Korea.