Discovery of a Janus Kinase Inhibitor Bearing a Highly Three-Dimensional Spiro Scaffold: JTE-052 (Delgocitinib) as a New Dermatological Agent to Treat Inflammatory Skin Disorders.
Noji, S., Hara, Y., Miura, T., Yamanaka, H., Maeda, K., Hori, A., Yamamoto, H., Obika, S., Inoue, M., Hase, Y., Orita, T., Doi, S., Adachi, T., Tanimoto, A., Oki, C., Kimoto, Y., Ogawa, Y., Negoro, T., Hashimoto, H., Shiozaki, M.(2020) J Med Chem 63: 7163-7185
- PubMed: 32511913
- DOI: https://doi.org/10.1021/acs.jmedchem.0c00450
- Primary Citation of Related Structures:
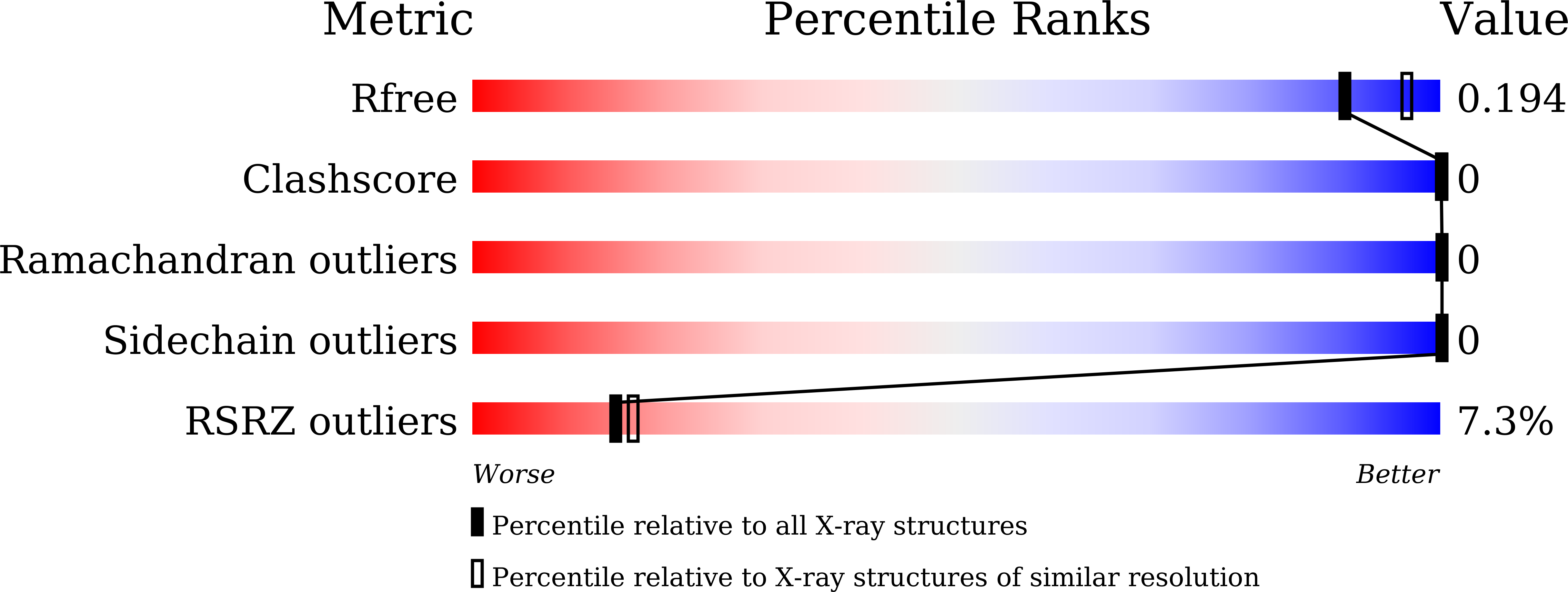
7C3N - PubMed Abstract:
Dermatologic disorders such as atopic dermatitis arise from genetic and environmental causes and are complex and multifactorial in nature. Among possible risk factors, aberrant immunological reactions are one of the leading etiologies. Immunosuppressive agents including topical steroids are common treatments for these disorders. Despite their reliability in clinical settings, topical steroids display side effects, typified by skin thinning. Accordingly, there is a need for alternate effective and well-tolerated therapies. As part of our efforts to investigate new immunomodulators, we have developed a series of JAK inhibitors, which incorporate novel three-dimensional spiro motifs and unexpectedly possess both excellent physicochemical properties and antidermatitis efficacy in the animal models. One of these compounds, JTE-052 ( ent - 60 ), also known as delgocitinib, has been shown to be effective and well-tolerated in human clinical trials and has recently been approved in Japan for the treatment of atopic dermatitis as the first drug among Janus kinase inhibitors.
Organizational Affiliation:
Chemical Research Laboratories, Central Pharmaceutical Research Institute, Japan Tobacco Inc., 1-1 Murasaki-cho, Takatsuki, Osaka 569-1125, Japan.