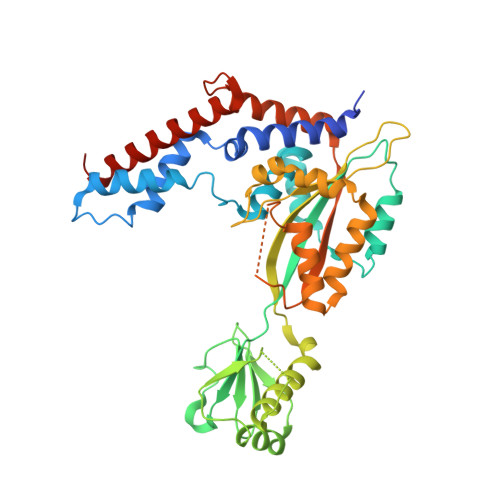
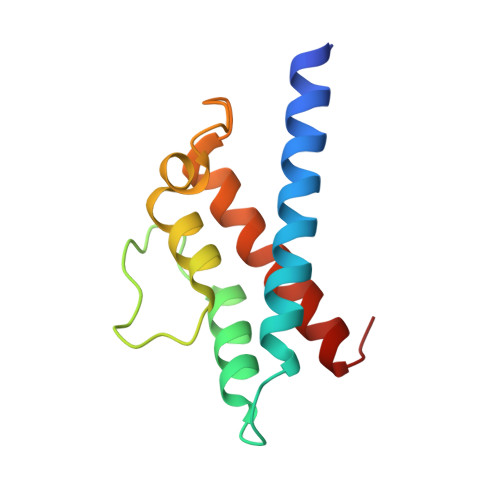
Insights into catalysis and regulation of non-canonical ubiquitination and deubiquitination by bacterial deamidase effectors.
Wang, Y., Zhan, Q., Wang, X., Li, P., Liu, S., Gao, G., Gao, P.(2020) Nat Commun 11: 2751-2751
- PubMed: 32488130
- DOI: https://doi.org/10.1038/s41467-020-16587-w
- Primary Citation of Related Structures:
7BXF, 7BXG, 7BXH - PubMed Abstract:
The bacterial effector MavC catalyzes non-canonical ubiquitination of host E2 enzyme UBE2N without engaging any of the conventional ubiquitination machinery, thereby abolishing UBE2N's function in forming K63-linked ubiquitin (Ub) chains and dampening NF-кB signaling. We now report the structures of MavC in complex with conjugated UBE2N~Ub and an inhibitor protein Lpg2149, as well as the structure of its ortholog, MvcA, bound to Lpg2149. Recognition of UBE2N and Ub depends on several unique features of MavC, which explains the inability of MvcA to catalyze ubiquitination. Unexpectedly, MavC and MvcA also possess deubiquitinase activity against MavC-mediated ubiquitination, highlighting MavC as a unique enzyme possessing deamidation, ubiquitination, and deubiquitination activities. Further, Lpg2149 directly binds and inhibits both MavC and MvcA by disrupting the interactions between enzymes and Ub. These results provide detailed insights into catalysis and regulation of MavC-type enzymes and the molecular mechanisms of this non-canonical ubiquitination machinery.
Organizational Affiliation:
CAS Key Laboratory of Infection and Immunity, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing, 100101, China.