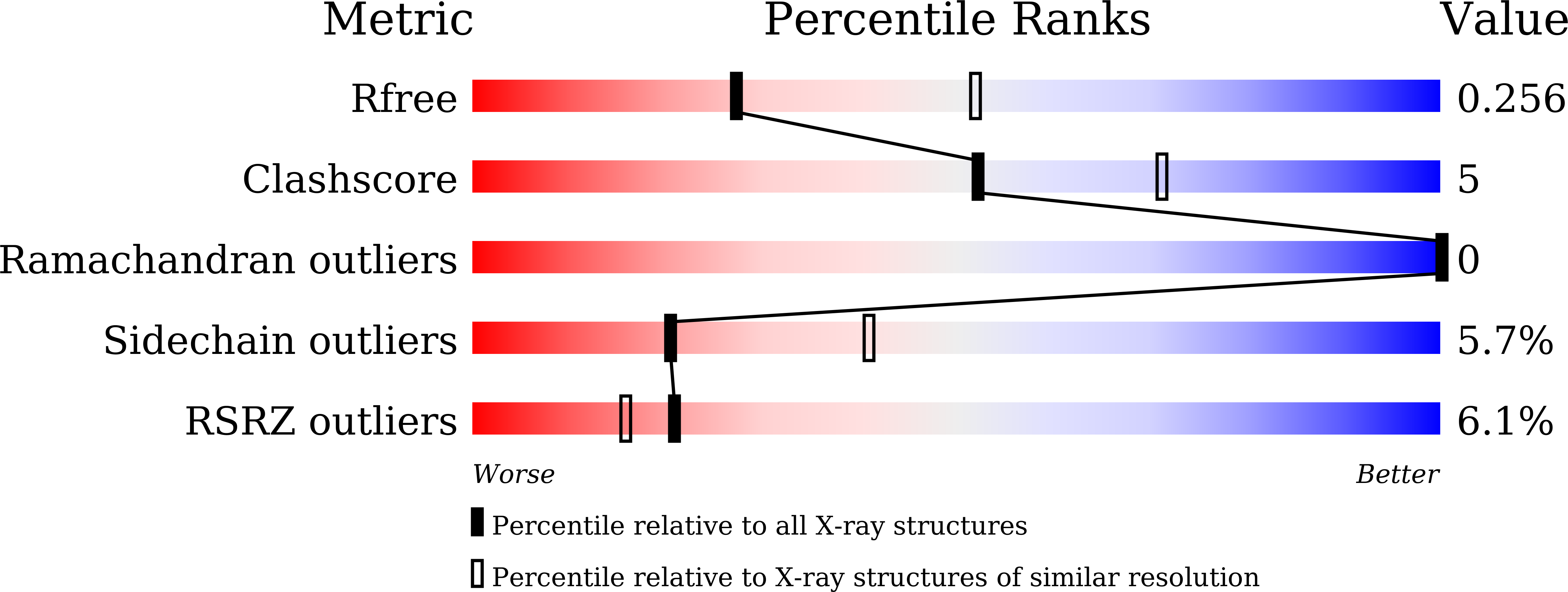
Molecular basis for the functions of dominantly active Y35N and inactive D60K Rheb mutants in mTORC1 signaling.
Zhang, C., Liu, Y., Zhang, Y., Wang, X., Zhang, T., Ding, J.(2020) J Mol Cell Biol 12: 741-744
- PubMed: 32470140
- DOI: https://doi.org/10.1093/jmcb/mjaa025
- Primary Citation of Related Structures:
7BTA, 7BTC, 7BTD
Organizational Affiliation:
State Key Laboratory of Molecular Biology, Shanghai Institute of Biochemistry and Cell Biology, Center for Excellence in Molecular Cell Science, University of Chinese Academy of Sciences, Chinese Academy of Sciences, Shanghai 200031, China.