Structural insight into hormone-recognition and transmembrane signaling by the atrial natriuretic peptide receptor
Ogawa, H.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Atrial natriuretic peptide receptor 1 | 435 | Rattus norvegicus | Mutation(s): 0 Gene Names: Npr1 EC: 4.6.1.2 | 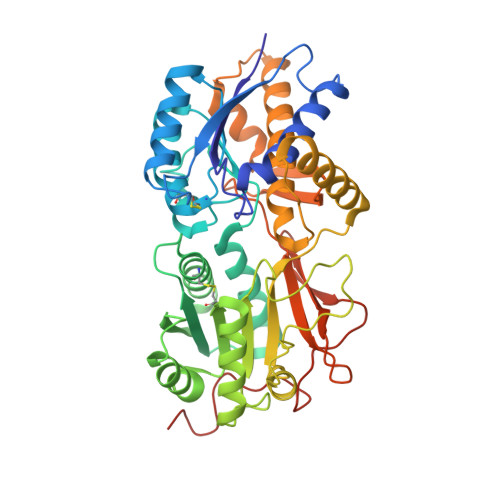 | |
UniProt | |||||
Find proteins for P18910 (Rattus norvegicus) Explore P18910 Go to UniProtKB: P18910 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P18910 | ||||
Glycosylation | |||||
| Glycosylation Sites: 2 | Go to GlyGen: P18910-1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Natriuretic peptides A | C [auth L] | 23 | Homo sapiens | Mutation(s): 0 |  |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P01160 (Homo sapiens) Explore P01160 Go to UniProtKB: P01160 | |||||
PHAROS: P01160 GTEx: ENSG00000175206 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P01160 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | D [auth C], F [auth E] | 3 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G62182OO GlyCosmos: G62182OO GlyGen: G62182OO | |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | E [auth D], G [auth F] | 4 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G48068RF GlyCosmos: G48068RF GlyGen: G48068RF | |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| CL (Subject of Investigation/LOI) Query on CL | H [auth A], I [auth B] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 99.419 | α = 90 |
| b = 99.419 | β = 90 |
| c = 259.666 | γ = 120 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| MOLREP | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Japan Society for the Promotion of Science (JSPS) | Japan | JP22570110 |
| Japan Agency for Medical Research and Development (AMED) | Japan | JP17am0101080, JP18am0101080 and JP19am0101080 |