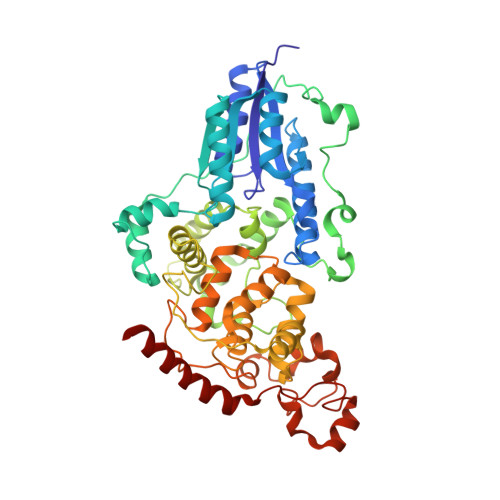
The three-dimensional structure of Drosophila melanogaster (6-4) photolyase at room temperature.
Cellini, A., Yuan Wahlgren, W., Henry, L., Pandey, S., Ghosh, S., Castillon, L., Claesson, E., Takala, H., Kubel, J., Nimmrich, A., Kuznetsova, V., Nango, E., Iwata, S., Owada, S., Stojkovic, E.A., Schmidt, M., Ihalainen, J.A., Westenhoff, S.(2021) Acta Crystallogr D Struct Biol 77: 1001-1009
- PubMed: 34342273
- DOI: https://doi.org/10.1107/S2059798321005830
- Primary Citation of Related Structures:
7AYV, 7AZT - PubMed Abstract:
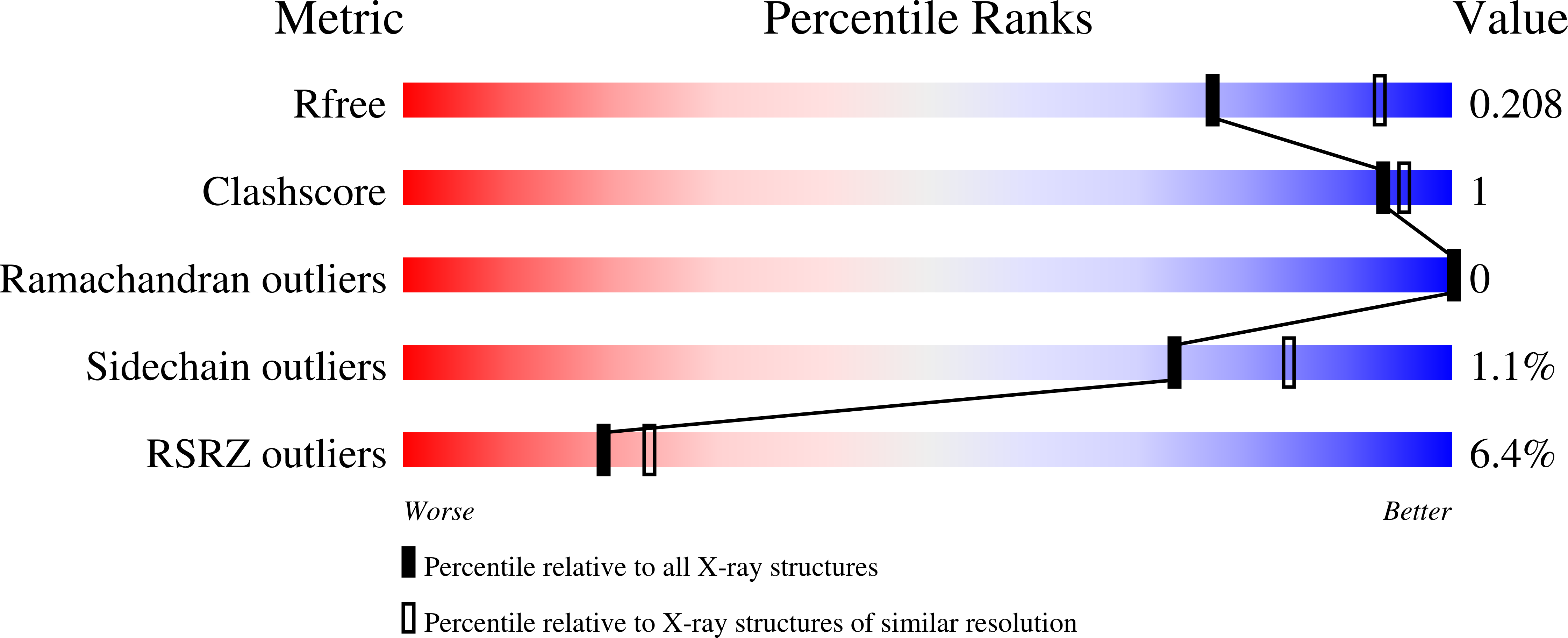
(6-4) photolyases are flavoproteins that belong to the photolyase/cryptochrome family. Their function is to repair DNA lesions using visible light. Here, crystal structures of Drosophila melanogaster (6-4) photolyase [Dm(6-4)photolyase] at room and cryogenic temperatures are reported. The room-temperature structure was solved to 2.27 Å resolution and was obtained by serial femtosecond crystallography (SFX) using an X-ray free-electron laser. The crystallization and preparation conditions are also reported. The cryogenic structure was solved to 1.79 Å resolution using conventional X-ray crystallography. The structures agree with each other, indicating that the structural information obtained from crystallography at cryogenic temperature also applies at room temperature. Furthermore, UV-Vis absorption spectroscopy confirms that Dm(6-4)photolyase is photoactive in the crystals, giving a green light to time-resolved SFX studies on the protein, which can reveal the structural mechanism of the photoactivated protein in DNA repair.
Organizational Affiliation:
Department of Chemistry and Molecular Biology, University of Gothenburg, Box 462, 405 30 Gothenburg, Sweden.