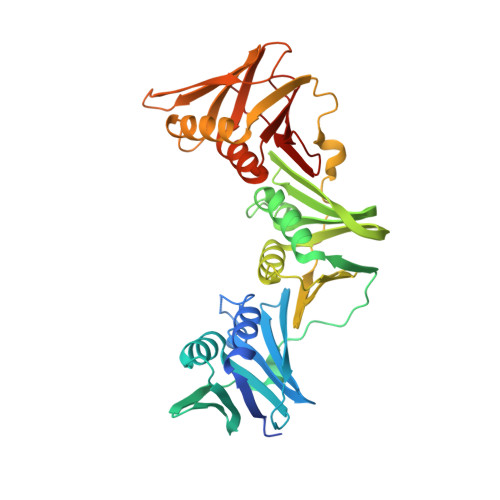
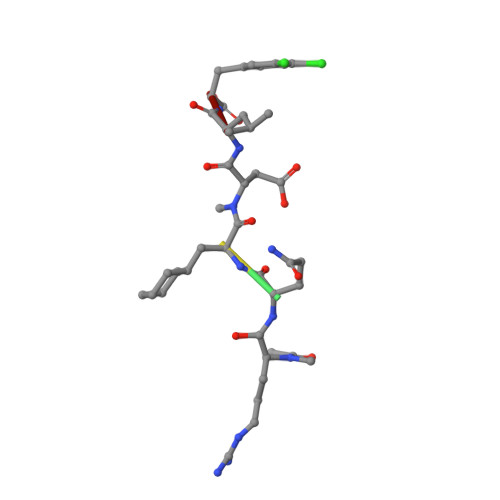
Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
Monsarrat, C., Compain, G., Andre, C., Engilberge, S., Martiel, I., Olieric, V., Wolff, P., Brillet, K., Landolfo, M., Silva da Veiga, C., Wagner, J., Guichard, G., Burnouf, D.Y.(2021) J Med Chem 64: 17063-17078
- PubMed: 34806883
- DOI: https://doi.org/10.1021/acs.jmedchem.1c00918
- Primary Citation of Related Structures:
7AZ5, 7AZ6, 7AZ7, 7AZ8, 7AZC, 7AZD, 7AZE, 7AZF, 7AZG, 7AZK, 7AZL - PubMed Abstract:
The bacterial DNA sliding clamp (SC), or replication processivity factor, is a promising target for the development of novel antibiotics. We report a structure-activity relationship study of a new series of peptides interacting within the Escherichia coli SC ( Ec SC) binding pocket. Various modifications were explored including N-alkylation of the peptide bonds, extension of the N-terminal moiety, and introduction of hydrophobic and constrained residues at the C-terminus. In each category, single modifications were identified that increased affinity to Ec SC. A combination of such modifications yielded in several cases to a substantially increased affinity compared to the parent peptides with K d in the range of 30-80 nM. X-ray structure analysis of 11 peptide/ Ec SC co-crystals revealed new interactions at the peptide-protein interface (i.e., stacking interactions, hydrogen bonds, and hydrophobic contacts) that can account for the improved binding. Several compounds among the best binders were also found to be more effective in inhibiting SC-dependent DNA synthesis.
Organizational Affiliation:
Université de Bordeaux, CNRS, Bordeaux INP, CBMN, UMR 5248, Institut Européen de Chimie et Biologie, 2 rue Robert Escarpit, F-33607 Pessac, France.