HIV-1 Integrase Inhibitors with Modifications That Affect Their Potencies against Drug Resistant Integrase Mutants.
Smith, S.J., Zhao, X.Z., Passos, D.O., Pye, V.E., Cherepanov, P., Lyumkis, D., Burke Jr., T.R., Hughes, S.H.(2021) ACS Infect Dis 7: 1469-1482
- PubMed: 33686850
- DOI: https://doi.org/10.1021/acsinfecdis.0c00819
- Primary Citation of Related Structures:
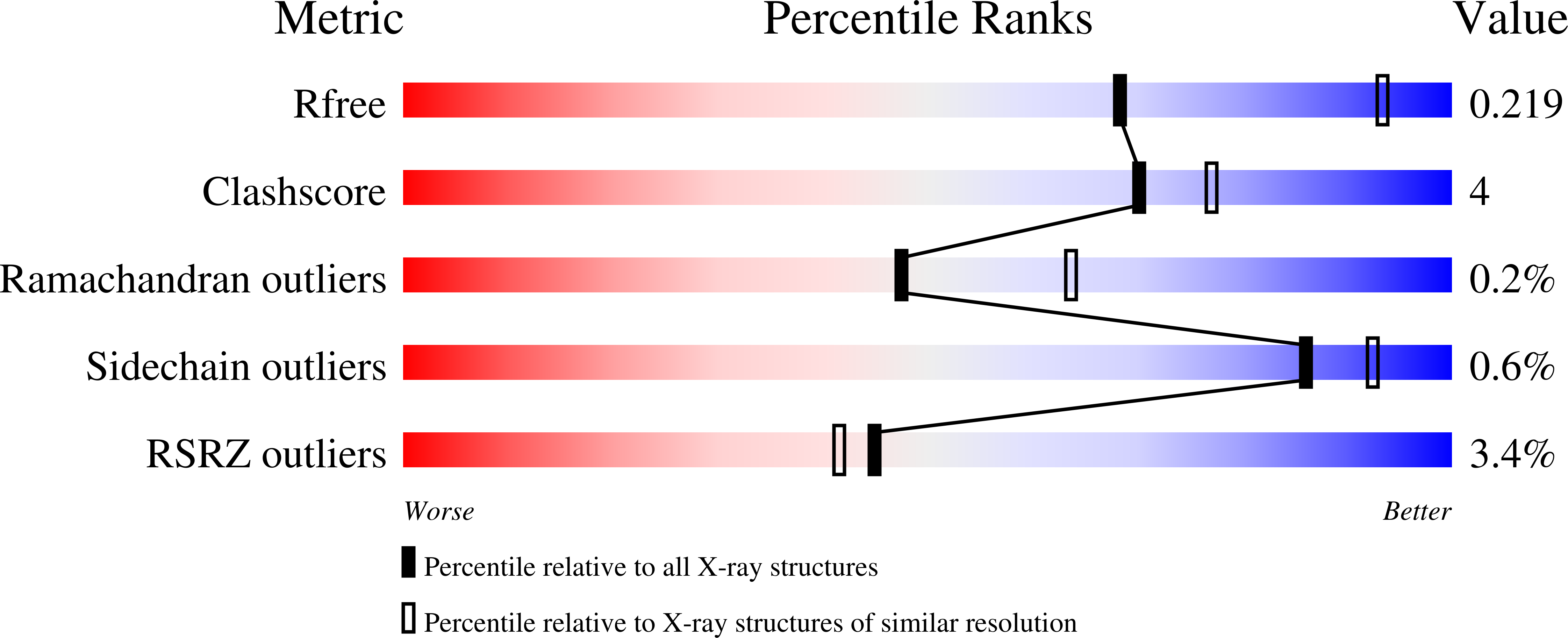
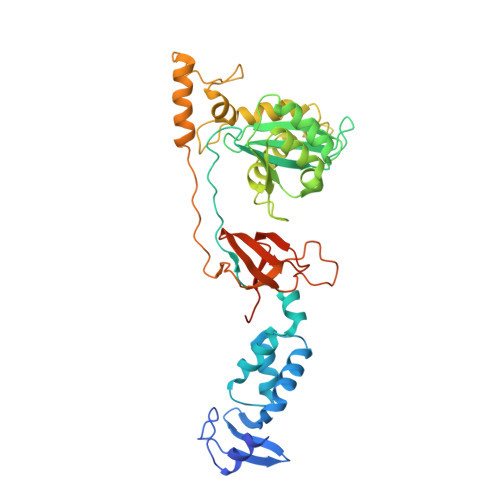
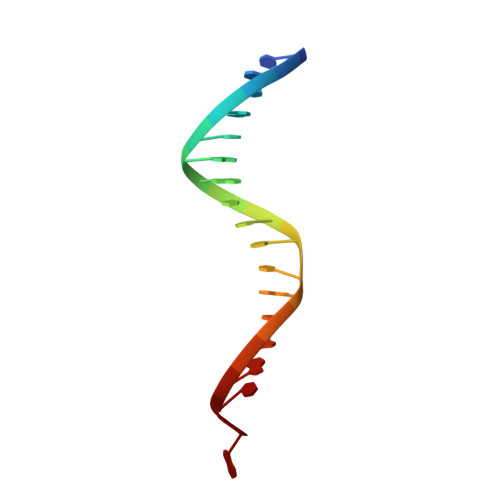
7ADU, 7ADV - PubMed Abstract:
Integrase strand transfer inhibitors (INSTIs) block the integration step of the retroviral lifecycle and are first-line drugs used for the treatment of HIV-1/AIDS. INSTIs have a polycyclic core with heteroatom triads, chelate the metal ions at the active site, and have a halobenzyl group that interacts with viral DNA attached to the core by a flexible linker. The most broadly effective INSTIs inhibit both wild-type (WT) integrase (IN) and a variety of well-known mutants. However, because there are mutations that reduce the potency of all of the available INSTIs, new and better compounds are needed. Models based on recent structures of HIV-1 and red-capped mangabey SIV INs suggest modifications in the INSTI structures that could enhance interactions with the 3'-terminal adenosine of the viral DNA, which could improve performance against INSTI resistant mutants. We designed and tested a series of INSTIs having modifications to their naphthyridine scaffold. One of the new compounds retained good potency against an expanded panel of HIV-1 IN mutants that we tested. Our results suggest the possibility of designing inhibitors that combine the best features of the existing compounds, which could provide additional efficacy against known HIV-1 IN mutants.
Organizational Affiliation:
HIV Dynamics and Replication Program, Center for Cancer Research, National Cancer Institute, Frederick, Maryland 21702, United States.