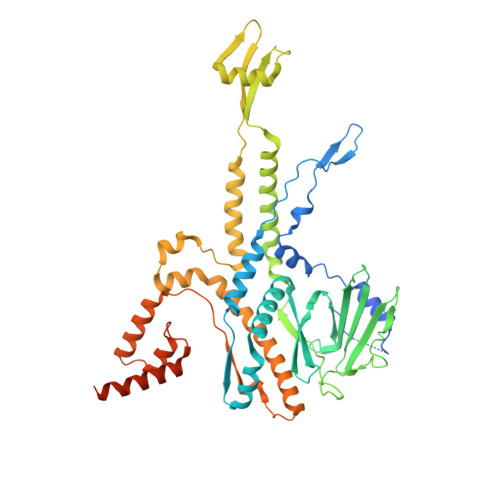
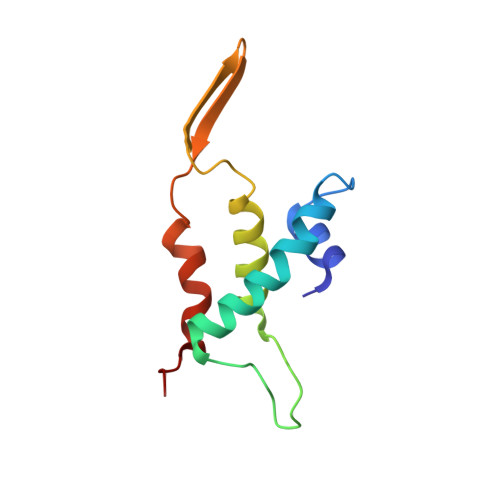
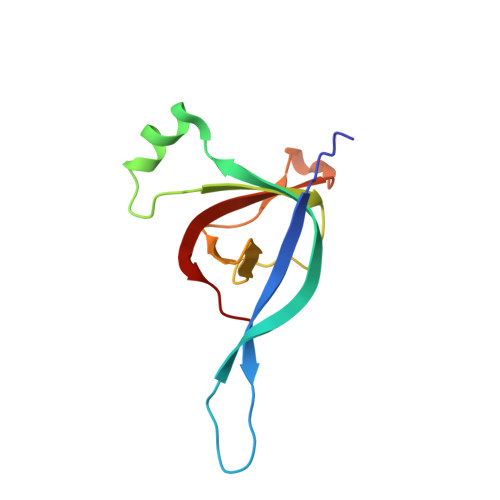
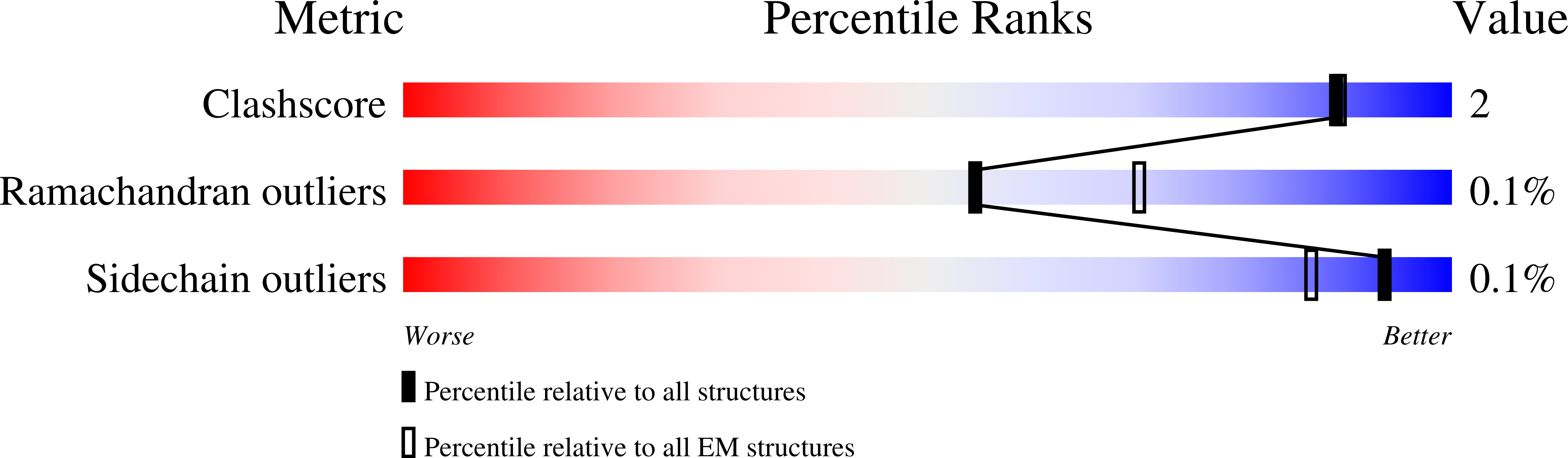CryoEM structure and assembly mechanism of a bacterial virus genome gatekeeper.
Orlov, I., Roche, S., Brasiles, S., Lukoyanova, N., Vaney, M.C., Tavares, P., Orlova, E.V.(2022) Nat Commun 13: 7283-7283
- PubMed: 36435855
- DOI: https://doi.org/10.1038/s41467-022-34999-8
- Primary Citation of Related Structures:
7Z4W - PubMed Abstract:
Numerous viruses package their dsDNA genome into preformed capsids through a portal gatekeeper that is subsequently closed. We report the structure of the DNA gatekeeper complex of bacteriophage SPP1 (gp6 12 gp15 12 gp16 6 ) in the post-DNA packaging state at 2.7 Å resolution obtained by single particle cryo-electron microscopy. Comparison of the native SPP1 complex with assembly-naïve structures of individual components uncovered the complex program of conformational changes leading to its assembly. After DNA packaging, gp15 binds via its C-terminus to the gp6 oligomer positioning gp15 subunits for oligomerization. Gp15 refolds its inner loops creating an intersubunit β-barrel that establishes different types of contacts with six gp16 subunits. Gp16 binding and oligomerization is accompanied by folding of helices that close the portal channel to keep the viral genome inside the capsid. This mechanism of assembly has broad functional and evolutionary implications for viruses of the prokaryotic tailed viruses-herpesviruses lineage.
Organizational Affiliation:
Centre for Integrative Biology (CBI), Department of Integrated Structural Biology, IGBMC, Université de Strasbourg, 67404, Illkirch, France.