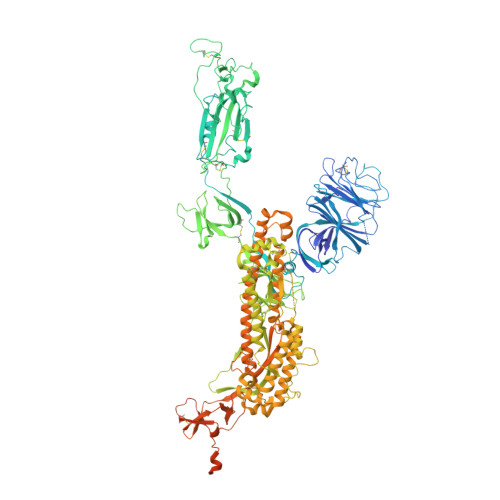
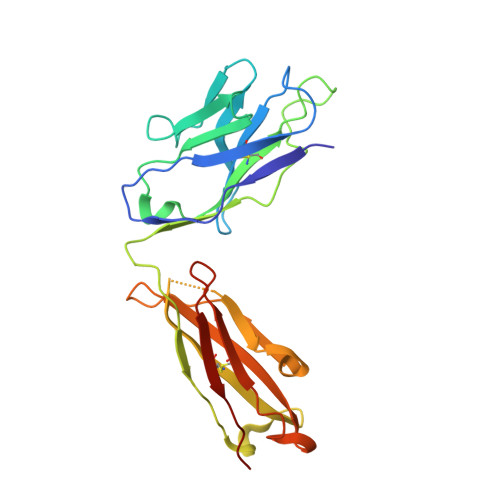
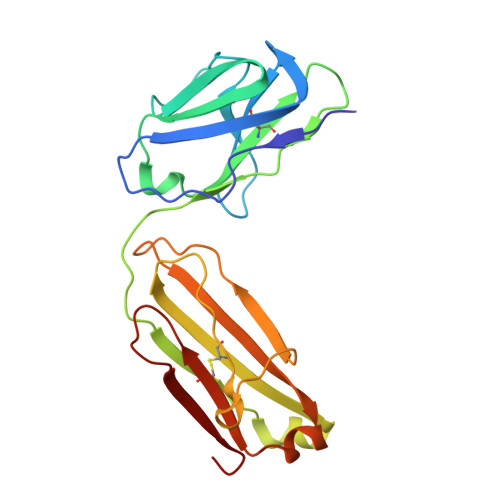
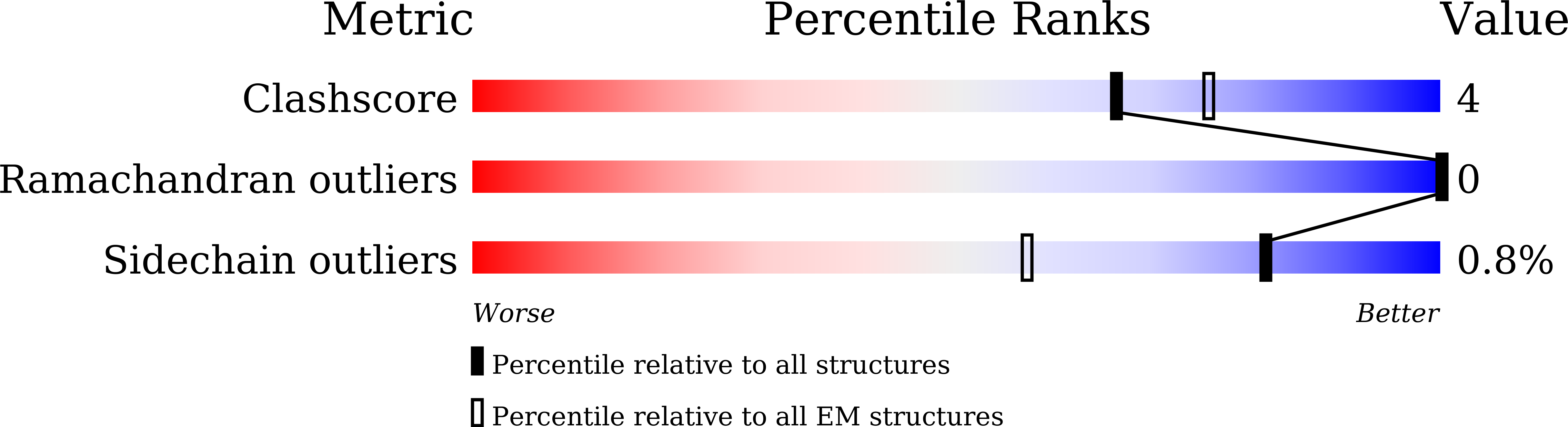Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.
Xu, Y., Wu, C., Cao, X., Gu, C., Liu, H., Jiang, M., Wang, X., Yuan, Q., Wu, K., Liu, J., Wang, D., He, X., Wang, X., Deng, S.J., Xu, H.E., Yin, W.(2022) Cell Res 32: 609-620
- PubMed: 35641567
- DOI: https://doi.org/10.1038/s41422-022-00672-4
- Primary Citation of Related Structures:
7XO4, 7XO5, 7XO6, 7XO7, 7XO8, 7XO9, 7XOA, 7XOB, 7XOC, 7XOD - PubMed Abstract:
The Omicron BA.2 variant has become a dominant infective strain worldwide. Receptor binding studies show that the Omicron BA.2 spike trimer exhibits 11-fold and 2-fold higher potency in binding to human ACE2 than the spike trimer from the wildtype (WT) and Omicron BA.1 strains. The structure of the BA.2 spike trimer complexed with human ACE2 reveals that all three receptor-binding domains (RBDs) in the spike trimer are in open conformation, ready for ACE2 binding, thus providing a basis for the increased infectivity of the BA.2 strain. JMB2002, a therapeutic antibody that was shown to efficiently inhibit Omicron BA.1, also shows potent neutralization activities against Omicron BA.2. In addition, both BA.1 and BA.2 spike trimers are able to bind to mouse ACE2 with high potency. In contrast, the WT spike trimer binds well to cat ACE2 but not to mouse ACE2. The structures of both BA.1 and BA.2 spike trimer bound to mouse ACE2 reveal the basis for their high affinity interactions. Together, these results suggest a possible evolution pathway for Omicron BA.1 and BA.2 variants via a human-cat-mouse-human circle, which could have important implications in establishing an effective strategy for combating SARS-CoV-2 viral infections.
Organizational Affiliation:
The CAS Key Laboratory of Receptor Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai, China.