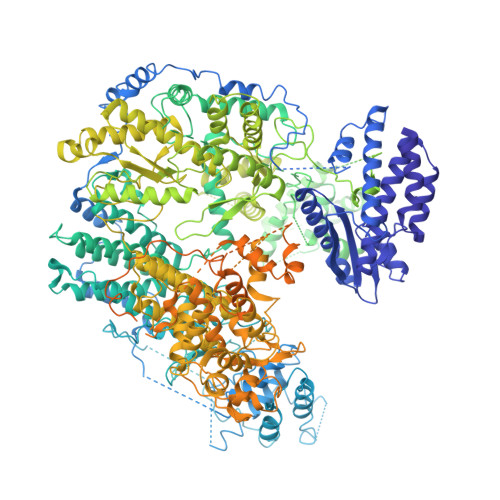
Structure basis for allosteric regulation of lymphocytic choriomeningitis virus polymerase function by Z matrix protein.
Liu, L., Wang, P., Liu, A., Zhang, L., Yan, L., Guo, Y., Xiao, G., Rao, Z., Lou, Z.(2023) Protein Cell 14: 703-707
- PubMed: 37038286
- DOI: https://doi.org/10.1093/procel/pwad018
- Primary Citation of Related Structures:
7X6S, 7X6V
Organizational Affiliation:
MOE Key Laboratory of Protein Science, School of Medicine, Tsinghua University, Beijing 100084, China.