Cryo-EM structure of SARS-CoV-2 S-Gamma variant (P.1) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, three ACE2-bound form conformation 2
Yang, T.J., Yu, P.Y., Chang, Y.C., Hsu, S.T.D.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Spike glycoprotein | 1,283 | Severe acute respiratory syndrome coronavirus 2 | Mutation(s): 16 Gene Names: S, 2 | 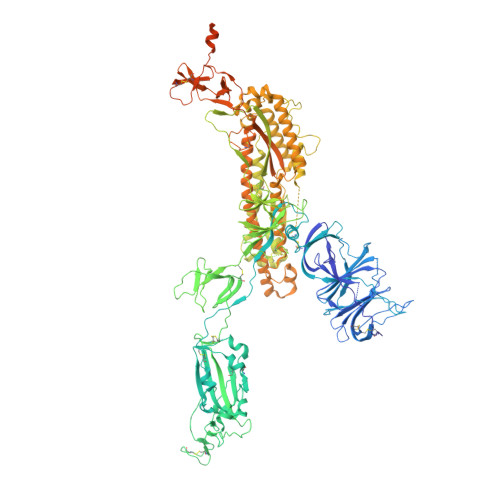 | |
UniProt | |||||
Find proteins for P0DTC2 (Severe acute respiratory syndrome coronavirus 2) Explore P0DTC2 Go to UniProtKB: P0DTC2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0DTC2 | ||||
Glycosylation | |||||
| Glycosylation Sites: 17 | Go to GlyGen: P0DTC2-1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Angiotensin-converting enzyme 2,Green fluorescent protein | 861 | Homo sapiens | Mutation(s): 0 Gene Names: ACE2, UNQ868/PRO1885 EC: 3.4.17.23 (PDB Primary Data), 3.4.17 (PDB Primary Data) | 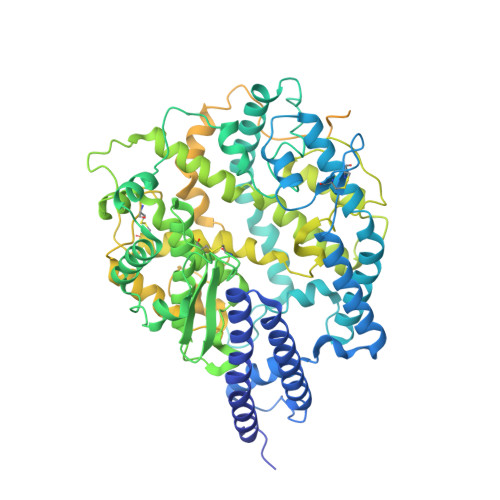 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for A0A059PIQ0 (Aequorea victoria) Explore A0A059PIQ0 Go to UniProtKB: A0A059PIQ0 | |||||
Find proteins for Q9BYF1 (Homo sapiens) Explore Q9BYF1 Go to UniProtKB: Q9BYF1 | |||||
PHAROS: Q9BYF1 GTEx: ENSG00000130234 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Groups | A0A059PIQ0Q9BYF1 | ||||
Glycosylation | |||||
| Glycosylation Sites: 6 | Go to GlyGen: Q9BYF1-1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | AA [auth a], BA [auth b], CA [auth c], DA [auth d], EA [auth e], AA [auth a], BA [auth b], CA [auth c], DA [auth d], EA [auth e], FA [auth f], G, GA [auth g], H, HA [auth h], I, IA [auth i], J, JA [auth j], K, L, M, N, O, P, Q, R, S, T, U, V, W, X, Y, Z | 2 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G42666HT GlyCosmos: G42666HT GlyGen: G42666HT | |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAG Query on NAG | AB [auth C] BB [auth C] CB [auth C] DB [auth C] EB [auth C] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| RECONSTRUCTION | cryoSPARC |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Ministry of Science and Technology (MoST, Taiwan) | Taiwan | 109-3114-Y-001-001 |
| Academia Sinica (Taiwan) | Taiwan | AS-CDA-109-L08 |
| Academia Sinica (Taiwan) | Taiwan | AS-CFII-108-110 |
| Academia Sinica (Taiwan) | Taiwan | AS-CFII108-111 |