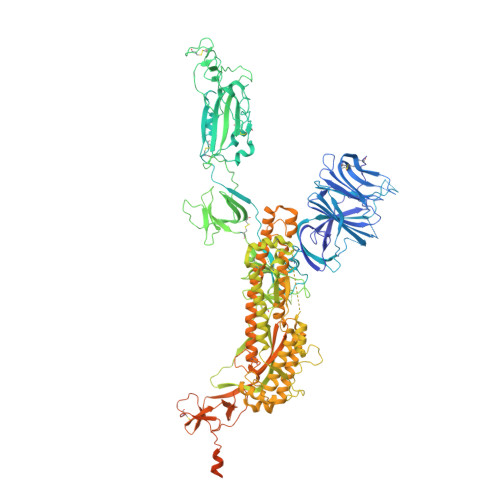
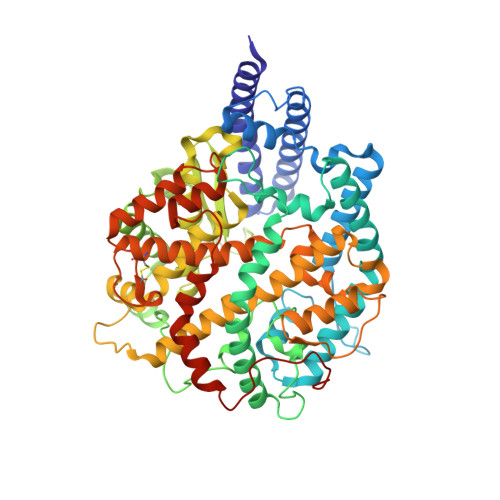
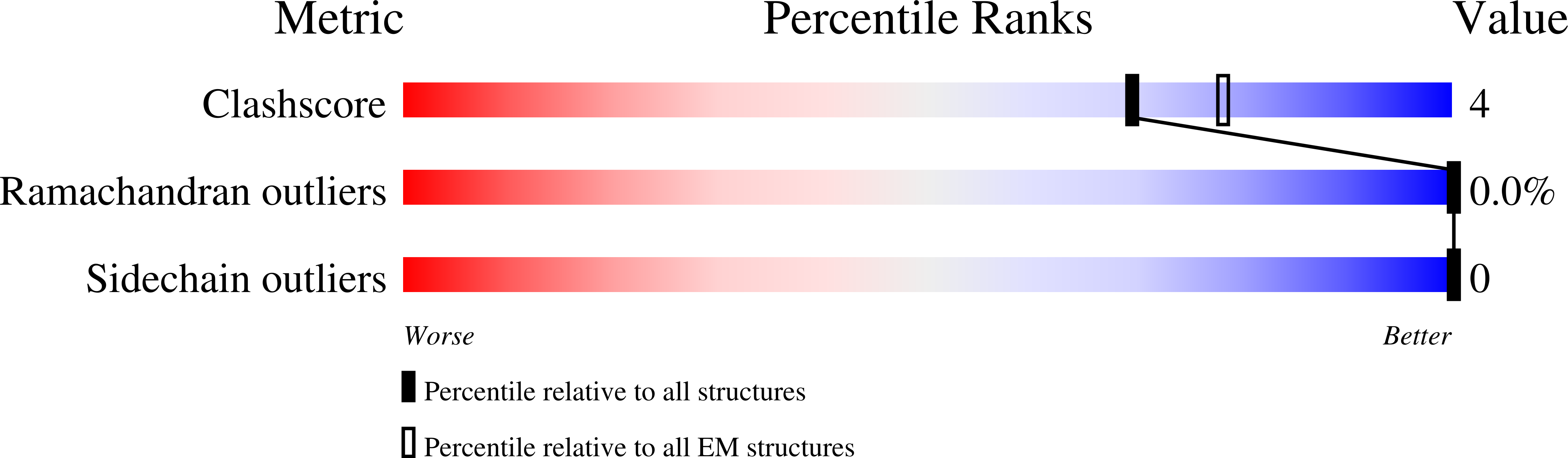SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein-ACE2 complex.
Mannar, D., Saville, J.W., Zhu, X., Srivastava, S.S., Berezuk, A.M., Tuttle, K.S., Marquez, A.C., Sekirov, I., Subramaniam, S.(2022) Science 375: 760-764
- PubMed: 35050643
- DOI: https://doi.org/10.1126/science.abn7760
- Primary Citation of Related Structures:
7T9J, 7T9K, 7T9L - PubMed Abstract:
The newly reported Omicron variant is poised to replace Delta as the most prevalent SARS-CoV-2 variant across the world. Cryo-EM structural analysis of the Omicron variant spike protein in complex with human ACE2 reveals new salt bridges and hydrogen bonds formed by mutated residues R493, S496 and R498 in the RBD with ACE2. These interactions appear to compensate for other Omicron mutations such as K417N known to reduce ACE2 binding affinity, resulting in similar biochemical ACE2 binding affinities for Delta and Omicron variants. Neutralization assays show that pseudoviruses displaying the Omicron spike protein exhibit increased antibody evasion. The increase in antibody evasion, together with retention of strong interactions at the ACE2 interface, thus represent important molecular features that likely contribute to the rapid spread of the Omicron variant.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, University of British Columbia, Vancouver, BC, Canada.