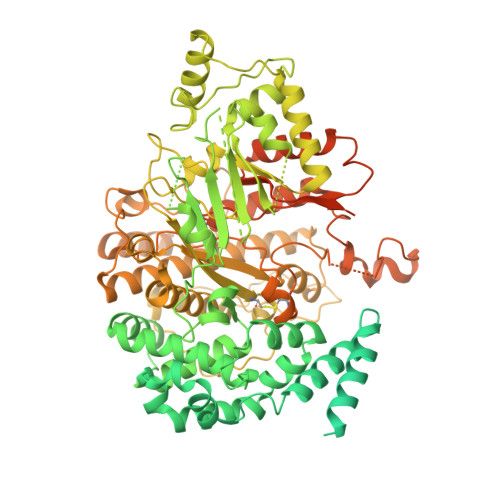
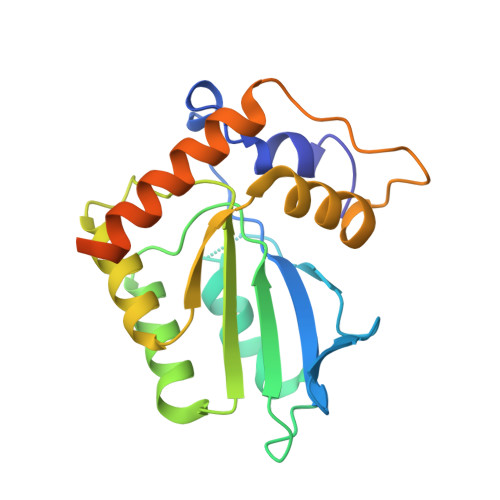
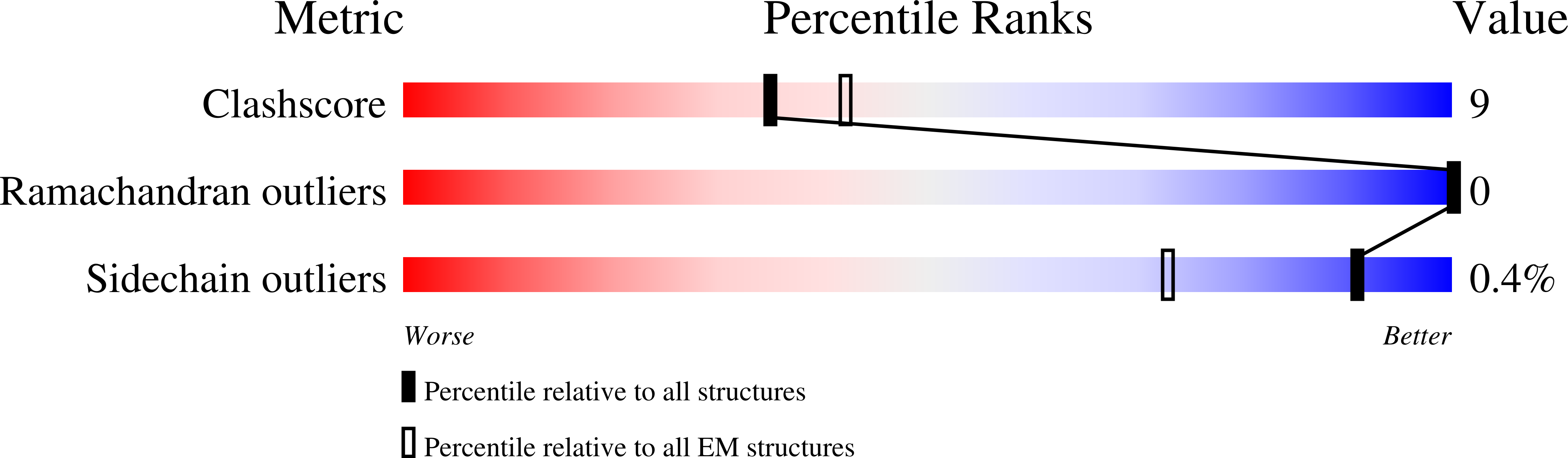Cryo-EM structure of the EBV ribonucleotide reductase BORF2 and mechanism of APOBEC3B inhibition.
Shaban, N.M., Yan, R., Shi, K., Moraes, S.N., Cheng, A.Z., Carpenter, M.A., McLellan, J.S., Yu, Z., Harris, R.S.(2022) Sci Adv 8: eabm2827-eabm2827
- PubMed: 35476445
- DOI: https://doi.org/10.1126/sciadv.abm2827
- Primary Citation of Related Structures:
7RW6 - PubMed Abstract:
Viruses use a plethora of mechanisms to evade immune responses. A recent example is neutralization of the nuclear DNA cytosine deaminase APOBEC3B by the Epstein-Barr virus (EBV) ribonucleotide reductase subunit BORF2. Cryo-EM studies of APOBEC3B-BORF2 complexes reveal a large >1000-Å 2 binding surface composed of multiple structural elements from each protein, which effectively blocks the APOBEC3B active site from accessing single-stranded DNA substrates. Evolutionary optimization is suggested by unique insertions in BORF2 absent from other ribonucleotide reductases and preferential binding to APOBEC3B relative to the highly related APOBEC3A and APOBEC3G enzymes. A molecular understanding of this pathogen-host interaction has potential to inform the development of drugs that block the interaction and liberate the natural antiviral activity of APOBEC3B. In addition, given a role for APOBEC3B in cancer mutagenesis, it may also be possible for information from the interaction to be used to develop DNA deaminase inhibitors.
Organizational Affiliation:
Department of Biochemistry, Molecular Biology, and Biophysics, Institute for Molecular Virology, Masonic Cancer Center, University of Minnesota, Minneapolis, MN 55455, USA.