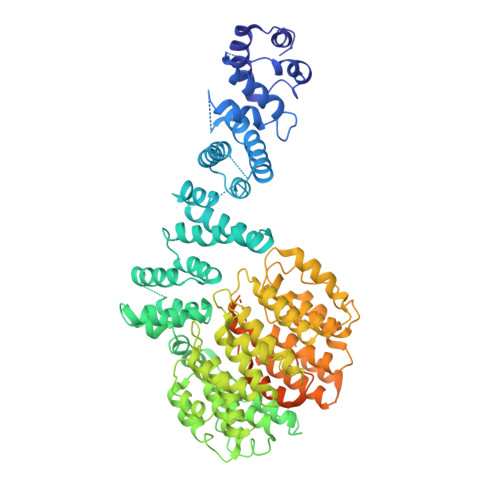
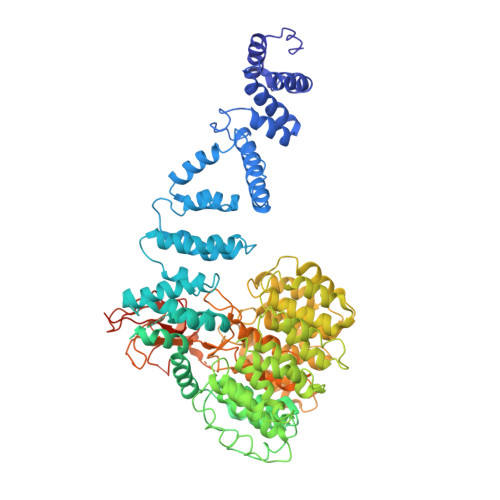
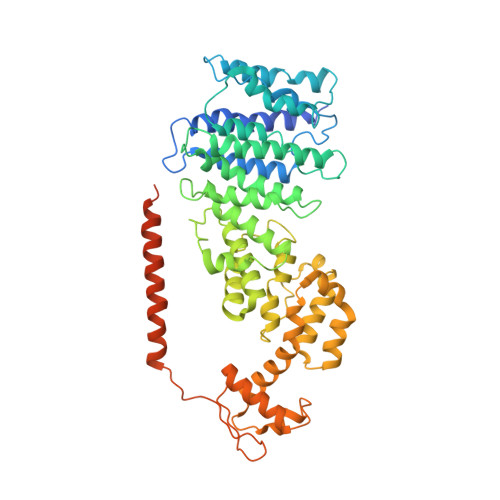
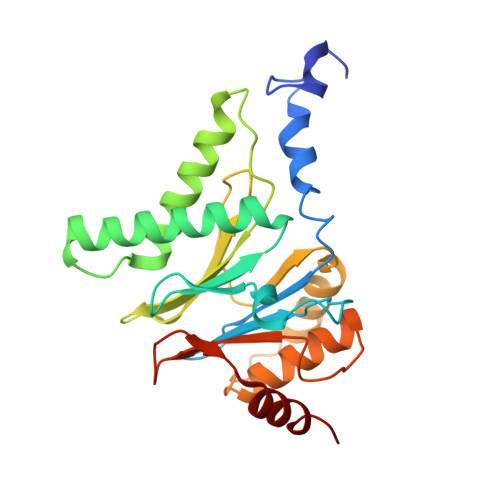
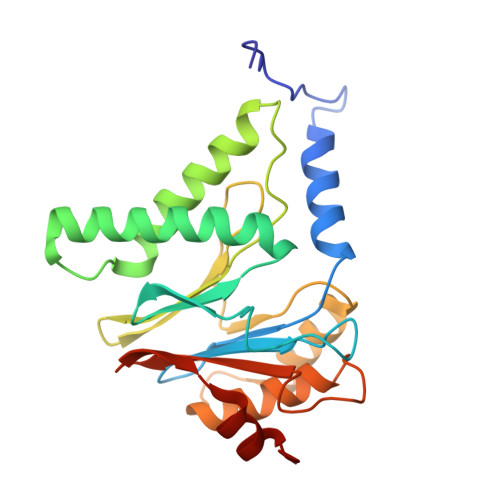
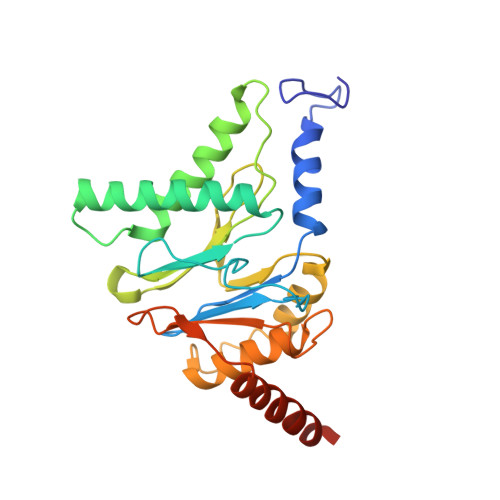
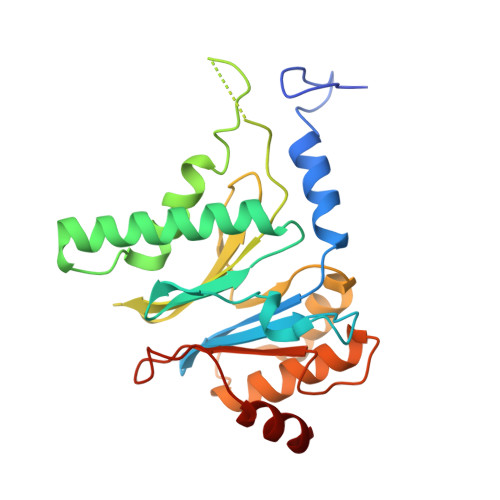
Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Lee, D., Zhu, Y., Colson, L., Wang, X., Chen, S., Tkacik, E., Huang, L., Ouyang, Q., Goldberg, A.L., Lu, Y.(2023) Mol Cell 83: 2959-2975.e7
- PubMed: 37595557
- DOI: https://doi.org/10.1016/j.molcel.2023.07.023
- Primary Citation of Related Structures:
7QXN, 7QXP, 7QXU, 7QXW, 7QXX, 7QY7, 7QYA, 7QYB - PubMed Abstract:
Various hormones, kinases, and stressors (fasting, heat shock) stimulate 26S proteasome activity. To understand how its capacity to degrade ubiquitylated proteins can increase, we studied mouse ZFAND5, which promotes protein degradation during muscle atrophy. Cryo-electron microscopy showed that ZFAND5 induces large conformational changes in the 19S regulatory particle. ZFAND5's AN1 Zn-finger domain interacts with the Rpt5 ATPase and its C terminus with Rpt1 ATPase and Rpn1, a ubiquitin-binding subunit. Upon proteasome binding, ZFAND5 widens the entrance of the substrate translocation channel, yet it associates only transiently with the proteasome. Dissociation of ZFAND5 then stimulates opening of the 20S proteasome gate. Using single-molecule microscopy, we showed that ZFAND5 binds ubiquitylated substrates, prolongs their association with proteasomes, and increases the likelihood that bound substrates undergo degradation, even though ZFAND5 dissociates before substrate deubiquitylation. These changes in proteasome conformation and reaction cycle can explain the accelerated degradation and suggest how other proteasome activators may stimulate proteolysis.
Organizational Affiliation:
Department of Cell Biology, Harvard Medical School, Boston, MA USA.