Viral peptide bound to chicken MHC-II molecule
Goryanin, A., Cook, A.G., Kaufman, J., Halabi, S.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| MHC class II alpha chain | 191 | Gallus gallus | Mutation(s): 0 Gene Names: B-LA | 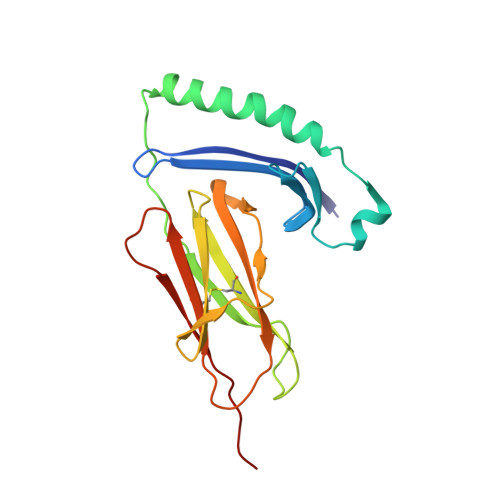 | |
UniProt | |||||
Find proteins for Q4U5Z6 (Gallus gallus) Explore Q4U5Z6 Go to UniProtKB: Q4U5Z6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q4U5Z6 | ||||
Glycosylation | |||||
| Glycosylation Sites: 1 | |||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 38 kDa phosphoprotein,MHC class II beta chain | 225 | Marek's disease herpesvirus (strain MD11/75C/R2), Gallus gallus This entity is chimeric | Mutation(s): 0 Gene Names: PP38 | 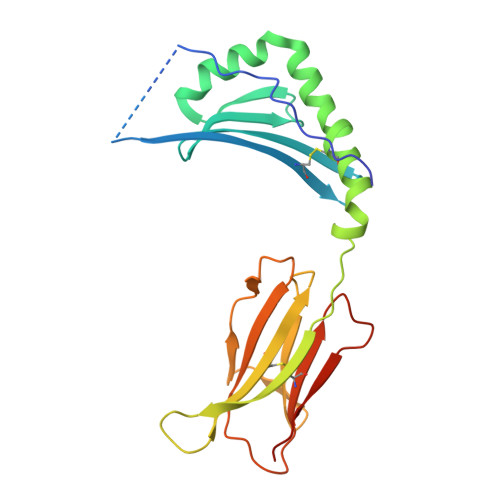 | |
UniProt | |||||
Find proteins for P68348 (Gallid herpesvirus 2 (strain MD11/75C/R2)) Explore P68348 Go to UniProtKB: P68348 | |||||
Find proteins for Q4U5Z9 (Gallus gallus) Explore Q4U5Z9 Go to UniProtKB: Q4U5Z9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Groups | Q4U5Z9P68348 | ||||
Glycosylation | |||||
| Glycosylation Sites: 1 | |||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAG Query on NAG | H [auth A], Q [auth C], U [auth D], X [auth F] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| PEG Query on PEG | L [auth A], O [auth B] | DI(HYDROXYETHYL)ETHER C4 H10 O3 MTHSVFCYNBDYFN-UHFFFAOYSA-N |  | ||
| GOL Query on GOL | I [auth A], K [auth A], M [auth B], T [auth C], W [auth E] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| ACT Query on ACT | J [auth A] N [auth B] P [auth B] R [auth C] S [auth C] | ACETATE ION C2 H3 O2 QTBSBXVTEAMEQO-UHFFFAOYSA-M |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 238.538 | α = 90 |
| b = 238.538 | β = 90 |
| c = 76.521 | γ = 120 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| Aimless | data scaling |
| PDB_EXTRACT | data extraction |
| xia2 | data reduction |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Wellcome Trust | United Kingdom | 110106/Z/15/Z |