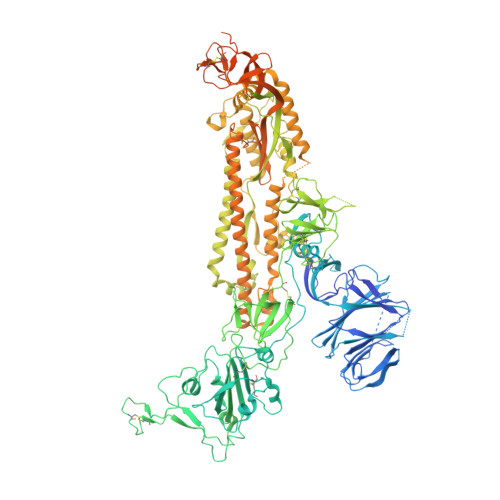
Structural insights in cell-type specific evolution of intra-host diversity by SARS-CoV-2.
Gupta, K., Toelzer, C., Williamson, M.K., Shoemark, D.K., Oliveira, A.S.F., Matthews, D.A., Almuqrin, A., Staufer, O., Yadav, S.K.N., Borucu, U., Garzoni, F., Fitzgerald, D., Spatz, J., Mulholland, A.J., Davidson, A.D., Schaffitzel, C., Berger, I.(2022) Nat Commun 13: 222-222
- PubMed: 35017512
- DOI: https://doi.org/10.1038/s41467-021-27881-6
- Primary Citation of Related Structures:
7OD3, 7ODL - PubMed Abstract:
As the global burden of SARS-CoV-2 infections escalates, so does the evolution of viral variants with increased transmissibility and pathology. In addition to this entrenched diversity, RNA viruses can also display genetic diversity within single infected hosts with co-existing viral variants evolving differently in distinct cell types. The BriSΔ variant, originally identified as a viral subpopulation from SARS-CoV-2 isolate hCoV-19/England/02/2020, comprises in the spike an eight amino-acid deletion encompassing a furin recognition motif and S1/S2 cleavage site. We elucidate the structure, function and molecular dynamics of this spike providing mechanistic insight into how the deletion correlates to viral cell tropism, ACE2 receptor binding and infectivity of this SARS-CoV-2 variant. Our results reveal long-range allosteric communication between functional domains that differ in the wild-type and the deletion variant and support a view of SARS-CoV-2 probing multiple evolutionary trajectories in distinct cell types within the same infected host.
Organizational Affiliation:
School of Biochemistry, University of Bristol, 1 Tankard's Close, Bristol, BS8 1TD, UK. kapil.gupta@bristol.ac.uk.