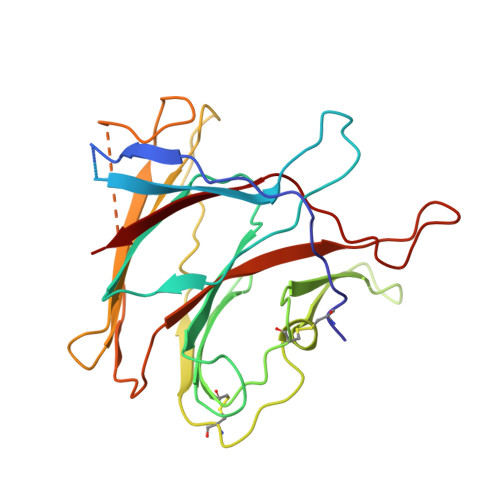
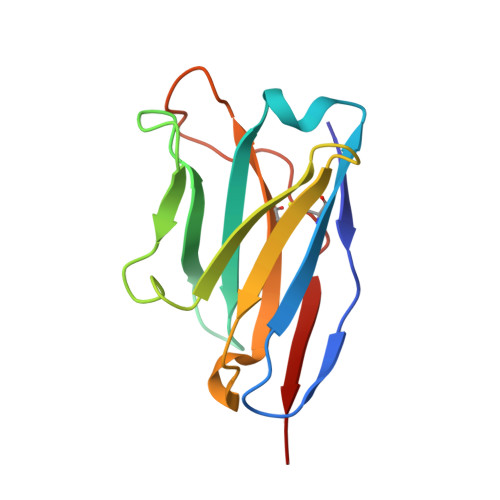
Prevalent, protective, and convergent IgG recognition of SARS-CoV-2 non-RBD spike epitopes.
Voss, W.N., Hou, Y.J., Johnson, N.V., Delidakis, G., Kim, J.E., Javanmardi, K., Horton, A.P., Bartzoka, F., Paresi, C.J., Tanno, Y., Chou, C.W., Abbasi, S.A., Pickens, W., George, K., Boutz, D.R., Towers, D.M., McDaniel, J.R., Billick, D., Goike, J., Rowe, L., Batra, D., Pohl, J., Lee, J., Gangappa, S., Sambhara, S., Gadush, M., Wang, N., Person, M.D., Iverson, B.L., Gollihar, J.D., Dye, J.M., Herbert, A.S., Finkelstein, I.J., Baric, R.S., McLellan, J.S., Georgiou, G., Lavinder, J.J., Ippolito, G.C.(2021) Science 372: 1108-1112
- PubMed: 33947773
- DOI: https://doi.org/10.1126/science.abg5268
- Primary Citation of Related Structures:
7M8J - PubMed Abstract:
The molecular composition and binding epitopes of the immunoglobulin G (IgG) antibodies that circulate in blood plasma after severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection are unknown. Proteomic deconvolution of the IgG repertoire to the spike glycoprotein in convalescent subjects revealed that the response is directed predominantly (>80%) against epitopes residing outside the receptor binding domain (RBD). In one subject, just four IgG lineages accounted for 93.5% of the response, including an amino (N)-terminal domain (NTD)-directed antibody that was protective against lethal viral challenge. Genetic, structural, and functional characterization of a multidonor class of "public" antibodies revealed an NTD epitope that is recurrently mutated among emerging SARS-CoV-2 variants of concern. These data show that "public" NTD-directed and other non-RBD plasma antibodies are prevalent and have implications for SARS-CoV-2 protection and antibody escape.
Organizational Affiliation:
Department of Molecular Biosciences, The University of Texas at Austin, Austin, TX, USA.