Crystal structure of putative NAD(P)H-flavin oxidoreductase from Haemophilus influenzae R2846
Maltseva, N., Kim, Y., Endres, M., Crofts, T., Joachimiak, A.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| NAD(P)H-dependent oxidoreductase | 223 | Haemophilus influenzae R2846 | Mutation(s): 0 Gene Names: CH638_05260 EC: 1 | 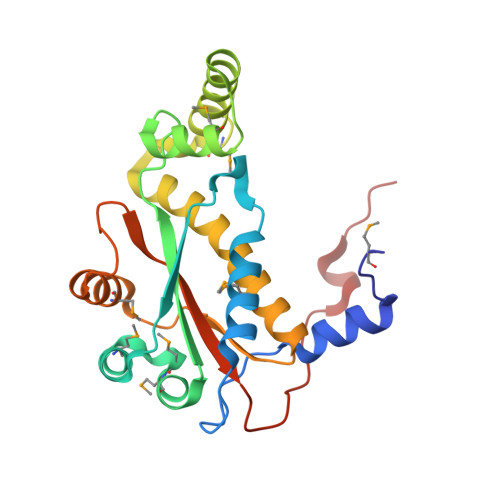 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 5 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| FMN (Subject of Investigation/LOI) Query on FMN | B [auth A] | FLAVIN MONONUCLEOTIDE C17 H21 N4 O9 P FVTCRASFADXXNN-SCRDCRAPSA-N |  | ||
| ACY Query on ACY | C [auth A] | ACETIC ACID C2 H4 O2 QTBSBXVTEAMEQO-UHFFFAOYSA-N |  | ||
| FMT Query on FMT | F [auth A], G [auth A] | FORMIC ACID C H2 O2 BDAGIHXWWSANSR-UHFFFAOYSA-N |  | ||
| CA Query on CA | D [auth A] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| CL Query on CL | E [auth A] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| MSE Query on MSE | A | L-PEPTIDE LINKING | C5 H11 N O2 Se |  | MET |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 53.1 | α = 90 |
| b = 84.838 | β = 115.831 |
| c = 53.977 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| PHENIX | refinement |
| HKL-3000 | data reduction |
| HKL-3000 | data scaling |
| HKL-3000 | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Human Genome Research Institute (NIH/NHGRI) | United States | -- |