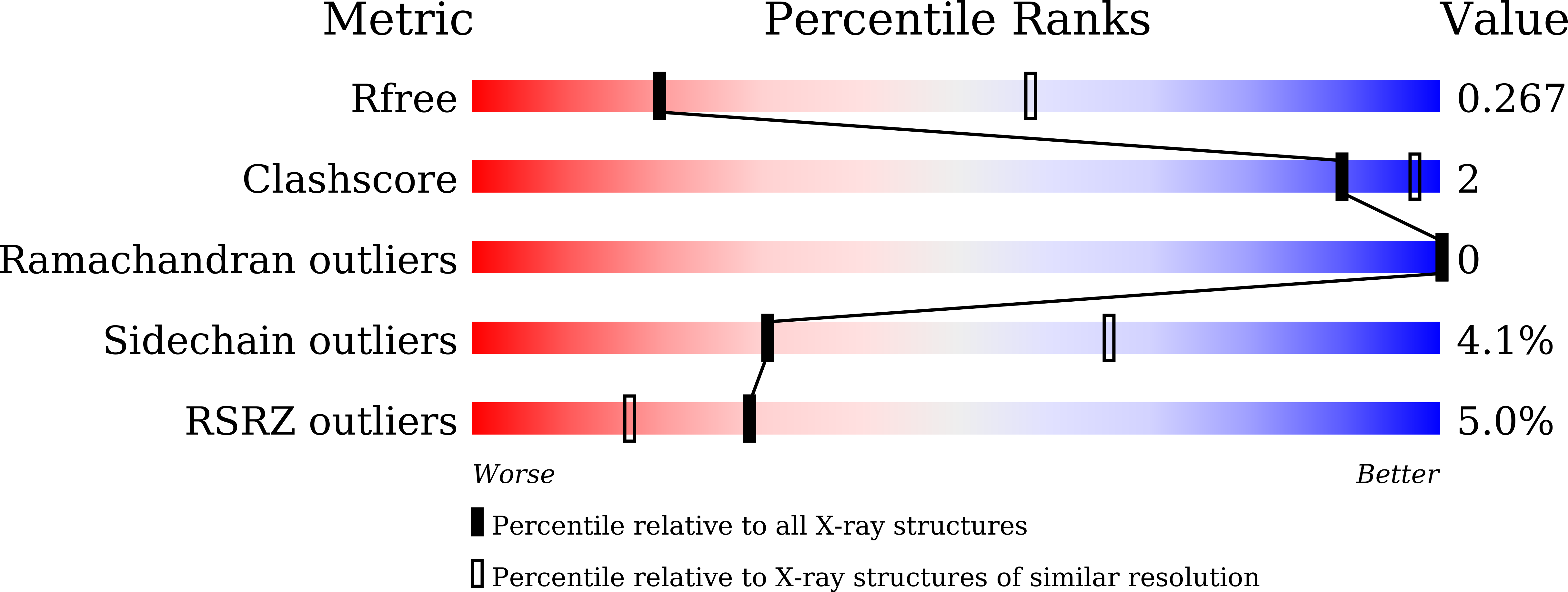
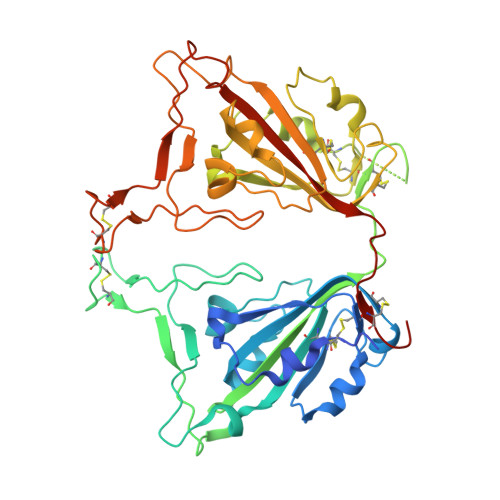
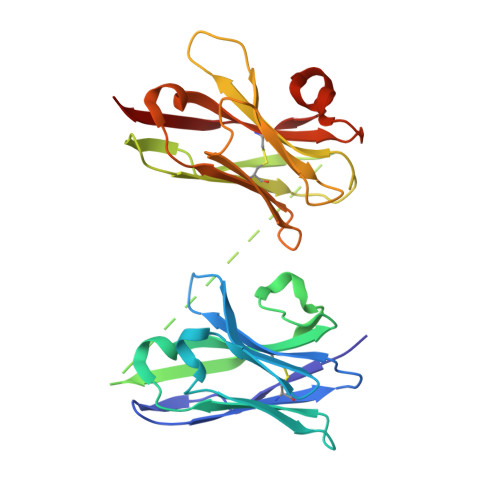
Hetero-bivalent nanobodies provide broad-spectrum protection against SARS-CoV-2 variants of concern including Omicron.
Ma, H., Zhang, X., Zheng, P., Dube, P.H., Zeng, W., Chen, S., Cheng, Q., Yang, Y., Wu, Y., Zhou, J., Hu, X., Xiang, Y., Zhang, H., Chiu, S., Jin, T.(2022) Cell Res 32: 831-842
- PubMed: 35906408
- DOI: https://doi.org/10.1038/s41422-022-00700-3
- Primary Citation of Related Structures:
7FH0, 7VOA - PubMed Abstract:
SARS-CoV-2 variants with adaptive mutations have continued to emerge, causing fresh waves of infection even amongst vaccinated population. The development of broad-spectrum antivirals is thus urgently needed. We previously developed two hetero-bivalent nanobodies (Nbs), aRBD-2-5 and aRBD-2-7, with potent neutralization activity against the wild-type (WT) Wuhan isolated SARS-CoV-2, by fusing aRBD-2 with aRBD-5 and aRBD-7, respectively. Here, we resolved the crystal structures of these Nbs in complex with the receptor-binding domain (RBD) of the spike protein, and found that aRBD-2 contacts with highly-conserved RBD residues and retains binding to the RBD of the Alpha, Beta, Gamma, Delta, Delta plus, Kappa, Lambda, Omicron BA.1, and BA.2 variants. In contrast, aRBD-5 and aRBD-7 bind to less-conserved RBD epitopes non-overlapping with the epitope of aRBD-2, and do not show apparent binding to the RBD of some variants. However, when fused with aRBD-2, they effectively enhance the overall binding affinity. Consistently, aRBD-2-5-Fc and aRBD-2-7-Fc potently neutralized all of the tested authentic or pseudotyped viruses, including WT, Alpha, Beta, Gamma, Delta, and Omicron BA.1, BA.1.1 and BA.2. Furthermore, aRBD-2-5-Fc provided prophylactic protection against the WT and mouse-adapted SARS-CoV-2 in mice, and conferred protection against the Omicron BA.1 variant in hamsters prophylactically and therapeutically, indicating that aRBD-2-5-Fc could potentially benefit the prevention and treatment of COVID-19 caused by the emerging variants of concern. Our strategy provides new solutions in the development of broad-spectrum therapeutic antibodies for COVID-19.
Organizational Affiliation:
Department of Pulmonary and Critical Care Medicine, The First Affiliated Hospital of USTC, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, Anhui, China.