Directed Evolution-Driven Increase of Structural Plasticity Is a Prerequisite for Binding the Complement Lectin Pathway Blocking MASP-Inhibitor Peptides.
Durvanger, Z., Boros, E., Nagy, Z.A., Hegedus, R., Megyeri, M., Dobo, J., Gal, P., Schlosser, G., Angyan, A.F., Gaspari, Z., Perczel, A., Harmat, V., Mezo, G., Menyhard, D.K., Pal, G.(2022) ACS Chem Biol 17: 969-986
- PubMed: 35378038
- DOI: https://doi.org/10.1021/acschembio.2c00114
- Primary Citation of Related Structures:
7ARX - PubMed Abstract:
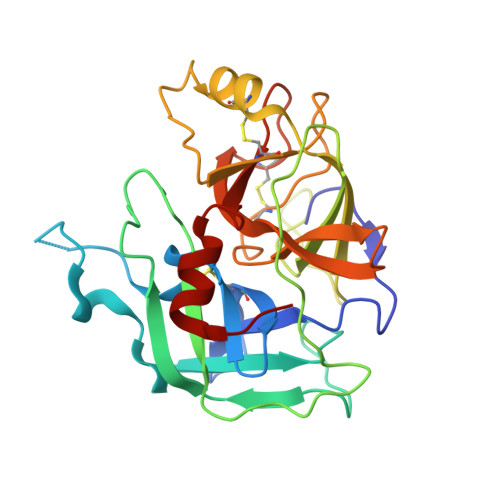
MASP-1 and MASP-2 are key activator proteases of the complement lectin pathway. The first specific mannose-binding lectin-associated serine protease (MASP) inhibitors had been developed from the 14-amino-acid sunflower trypsin inhibitor (SFTI) peptide by phage display, yielding SFTI-based MASP inhibitors, SFMIs. Here, we present the crystal structure of the MASP-1/SFMI1 complex that we analyzed in comparison to other existing MASP-1/2 structures. Rigidified backbone structure has long been accepted as a structural prerequisite for peptide inhibitors of proteases. We found that a hydrophobic cluster organized around the P2 Thr residue is essential for the structural stability of wild-type SFTI. We also found that the same P2 Thr prevents binding of the rigid SFTI-like peptides to the substrate-binding cleft of both MASPs as the cleft is partially blocked by large gatekeeper enzyme loops. Directed evolution removed this obstacle by replacing the P2 Thr with a Ser, providing the SFMIs with high-degree structural plasticity, which proved to be essential for MASP inhibition. To gain more insight into the structural criteria for SFMI-based MASP-2 inhibition, we systematically modified MASP-2-specific SFMI2 by capping its two termini and by replacing its disulfide bridge with varying length thioether linkers. By doing so, we also aimed to generate a versatile scaffold that is resistant to reducing environment and has increased stability in exopeptidase-containing biological environments. We found that the reduction-resistant disulfide-substituted l-2,3-diaminopropionic acid (Dap) variant possessed near-native potency. As MASP-2 is involved in the life-threatening thrombosis in COVID-19 patients, our synthetic, selective MASP-2 inhibitors could be relevant coronavirus drug candidates.
Organizational Affiliation:
Laboratory of Structural Chemistry and Biology, Institute of Chemistry, ELTE Eötvös Loránd University, Pázmány Péter sétány 1/A, H-1117 Budapest, Hungary.