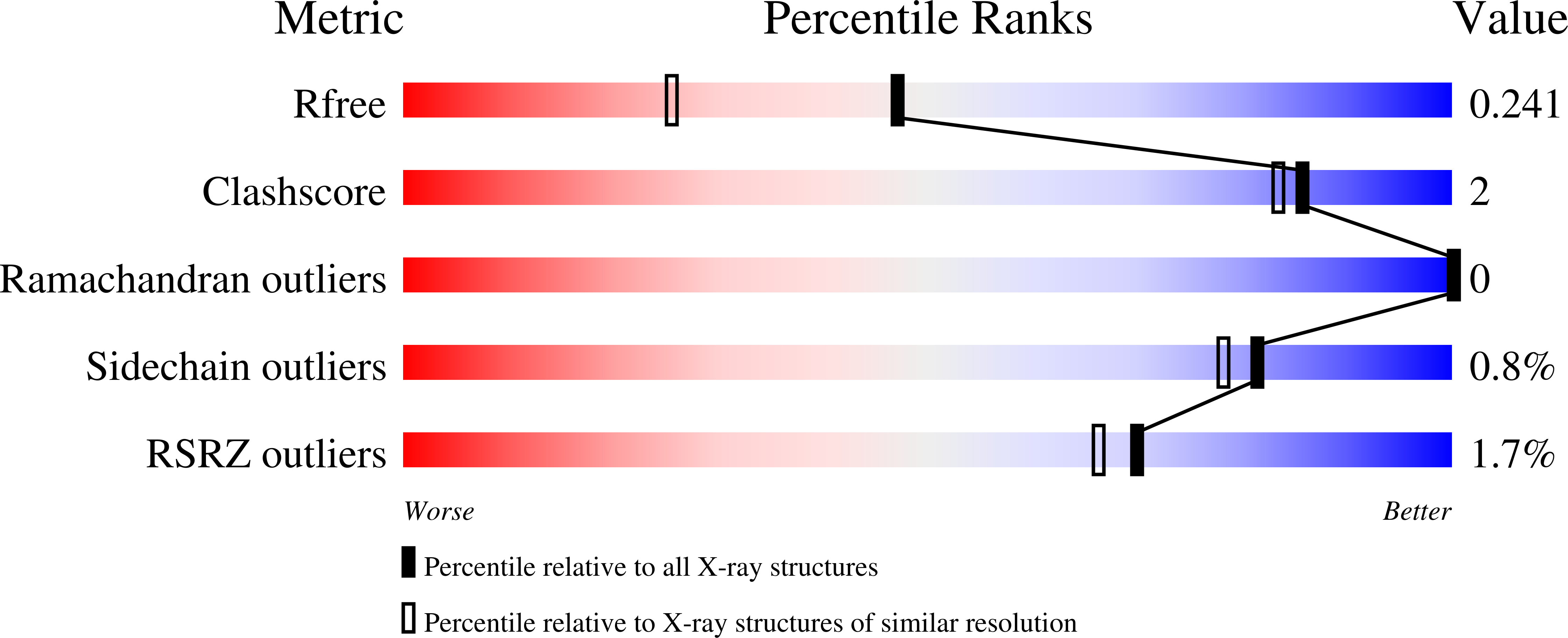
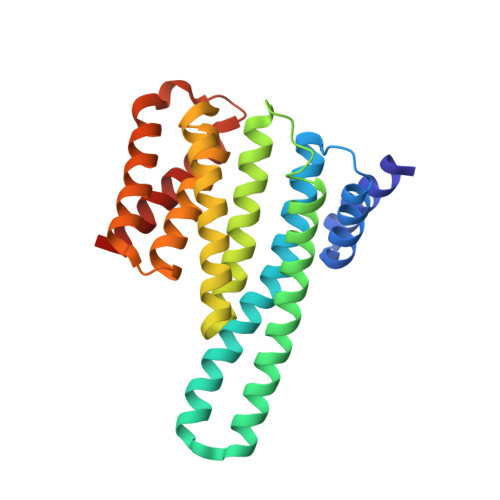
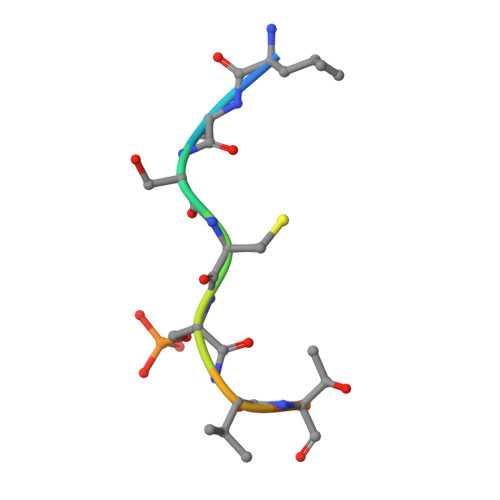
14-3-3-protein regulates Nedd4-2 by modulating interactions between HECT and WW domains.
Pohl, P., Joshi, R., Petrvalska, O., Obsil, T., Obsilova, V.(2021) Commun Biol 4: 899-899
- PubMed: 34294877
- DOI: https://doi.org/10.1038/s42003-021-02419-0
- Primary Citation of Related Structures:
6ZBT, 6ZC9, 7NMZ - PubMed Abstract:
Neural precursor cell expressed developmentally down-regulated 4 ligase (Nedd4-2) is an E3 ubiquitin ligase that targets proteins for ubiquitination and endocytosis, thereby regulating numerous ion channels, membrane receptors and tumor suppressors. Nedd4-2 activity is regulated by autoinhibition, calcium binding, oxidative stress, substrate binding, phosphorylation and 14-3-3 protein binding. However, the structural basis of 14-3-3-mediated Nedd4-2 regulation remains poorly understood. Here, we combined several techniques of integrative structural biology to characterize Nedd4-2 and its complex with 14-3-3. We demonstrate that phosphorylated Ser 342 and Ser 448 are the key residues that facilitate 14-3-3 protein binding to Nedd4-2 and that 14-3-3 protein binding induces a structural rearrangement of Nedd4-2 by inhibiting interactions between its structured domains. Overall, our findings provide the structural glimpse into the 14-3-3-mediated Nedd4-2 regulation and highlight the potential of the Nedd4-2:14-3-3 complex as a pharmacological target for Nedd4-2-associated diseases such as hypertension, epilepsy, kidney disease and cancer.
Organizational Affiliation:
Department of Structural Biology of Signaling Proteins, Division BIOCEV, Institute of Physiology of the Czech Academy of Sciences, Vestec, Czech Republic.