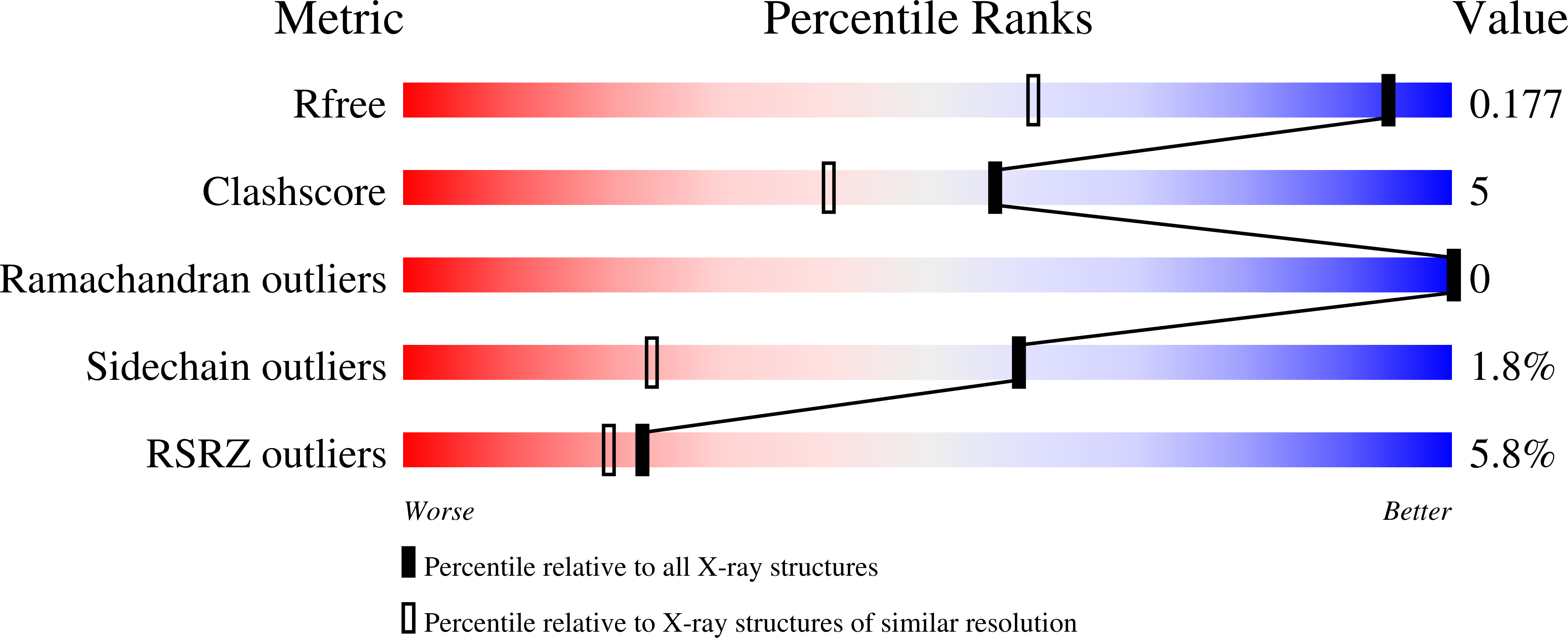
An unprecedented insight into the catalytic mechanism of copper nitrite reductase from atomic-resolution and damage-free structures.
Rose, S.L., Antonyuk, S.V., Sasaki, D., Yamashita, K., Hirata, K., Ueno, G., Ago, H., Eady, R.R., Tosha, T., Yamamoto, M., Hasnain, S.S.(2021) Sci Adv 7
- PubMed: 33523860
- DOI: https://doi.org/10.1126/sciadv.abd8523
- Primary Citation of Related Structures:
6ZAR, 6ZAS, 6ZAT, 6ZAU, 6ZAV, 6ZAW, 6ZAX - PubMed Abstract:
Copper-containing nitrite reductases (CuNiRs), encoded by nirK gene, are found in all kingdoms of life with only 5% of CuNiR denitrifiers having two or more copies of nirK Recently, we have identified two copies of nirK genes in several α-proteobacteria of the order Rhizobiales including Bradyrhizobium sp. ORS 375, encoding a four-domain heme-CuNiR and the usual two-domain CuNiR ( Br 2D NiR). Compared with two of the best-studied two-domain CuNiRs represented by the blue ( Ax NiR) and green ( Ac NiR) subclasses, Br 2D NiR, a blue CuNiR, shows a substantially lower catalytic efficiency despite a sequence identity of ~70%. Advanced synchrotron radiation and x-ray free-electron laser are used to obtain the most accurate (atomic resolution with unrestrained SHELX refinement) and damage-free (free from radiation-induced chemistry) structures, in as-isolated, substrate-bound, and product-bound states. This combination has shed light on the protonation states of essential catalytic residues, additional reaction intermediates, and how catalytic efficiency is modulated.
Organizational Affiliation:
Molecular Biophysics Group, Life Sciences Building and Institute of Systems, Molecular and Integrative Biology, Faculty of Health and Life Sciences, University of Liverpool, Liverpool L69 7ZB, UK.