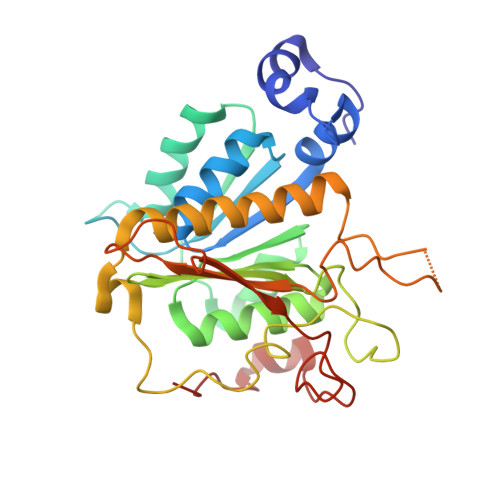
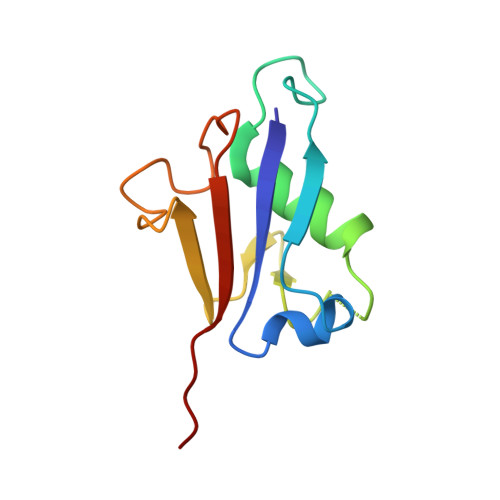
Molecular basis for the bifunctional Uba4-Urm1 sulfur-relay system in tRNA thiolation and ubiquitin-like conjugation.
Pabis, M., Termathe, M., Ravichandran, K.E., Kienast, S.D., Krutyholowa, R., Sokolowski, M., Jankowska, U., Grudnik, P., Leidel, S.A., Glatt, S.(2020) EMBO J 39: e105087-e105087
- PubMed: 32901956
- DOI: https://doi.org/10.15252/embj.2020105087
- Primary Citation of Related Structures:
6YUB, 6YUC, 6Z6S - PubMed Abstract:
The chemical modification of tRNA bases by sulfur is crucial to tune translation and to optimize protein synthesis. In eukaryotes, the ubiquitin-related modifier 1 (Urm1) pathway is responsible for the synthesis of 2-thiolated wobble uridine (U 34 ). During the key step of the modification cascade, the E1-like activating enzyme ubiquitin-like protein activator 4 (Uba4) first adenylates and thiocarboxylates the C-terminus of its substrate Urm1. Subsequently, activated thiocarboxylated Urm1 (Urm1-COSH) can serve as a sulfur donor for specific tRNA thiolases or participate in ubiquitin-like conjugation reactions. Structural and mechanistic details of Uba4 and Urm1 have remained elusive but are key to understand the evolutionary branch point between ubiquitin-like proteins (UBL) and sulfur-relay systems. Here, we report the crystal structures of full-length Uba4 and its heterodimeric complex with its substrate Urm1. We show how the two domains of Uba4 orchestrate recognition, binding, and thiocarboxylation of the C-terminus of Urm1. Finally, we uncover how the catalytic domains of Uba4 communicate efficiently during the reaction cycle and identify a mechanism that enables Uba4 to protect itself against self-conjugation with its own product, namely activated Urm1-COSH.
Organizational Affiliation:
Malopolska Centre of Biotechnology (MCB), Jagiellonian University, Krakow, Poland.