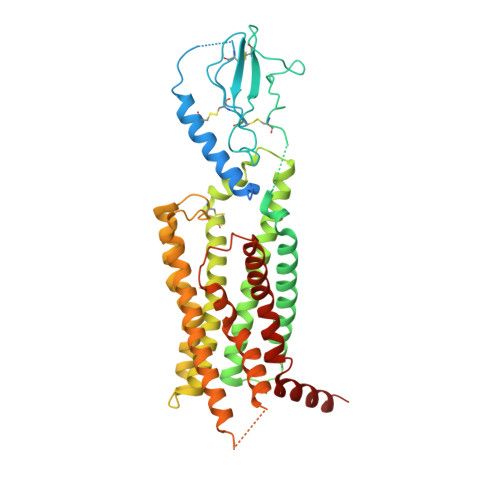
Structural basis for GLP-1 receptor activation by LY3502970, an orally active nonpeptide agonist.
Kawai, T., Sun, B., Yoshino, H., Feng, D., Suzuki, Y., Fukazawa, M., Nagao, S., Wainscott, D.B., Showalter, A.D., Droz, B.A., Kobilka, T.S., Coghlan, M.P., Willard, F.S., Kawabe, Y., Kobilka, B.K., Sloop, K.W.(2020) Proc Natl Acad Sci U S A 117: 29959-29967
- PubMed: 33177239
- DOI: https://doi.org/10.1073/pnas.2014879117
- Primary Citation of Related Structures:
6XOX - PubMed Abstract:
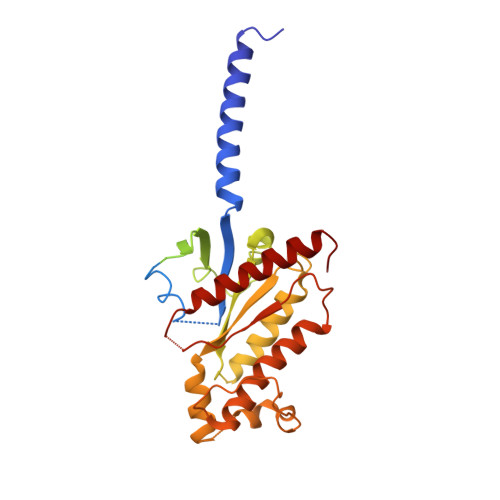
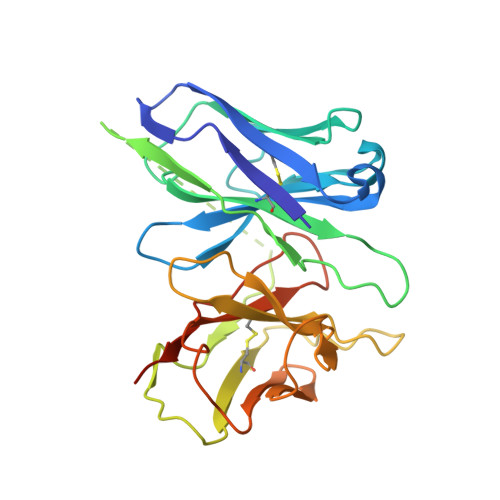
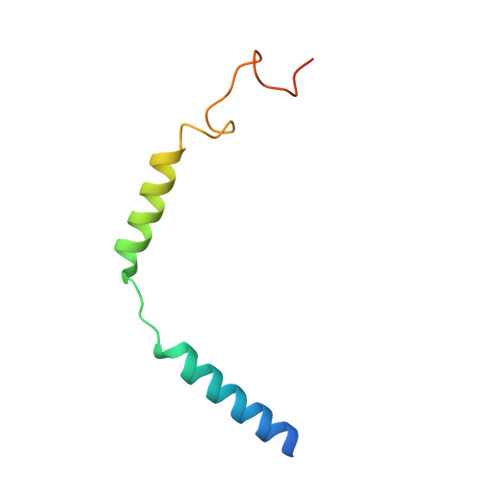
Glucagon-like peptide-1 receptor (GLP-1R) agonists are efficacious antidiabetic medications that work by enhancing glucose-dependent insulin secretion and improving energy balance. Currently approved GLP-1R agonists are peptide based, and it has proven difficult to obtain small-molecule activators possessing optimal pharmaceutical properties. We report the discovery and mechanism of action of LY3502970 (OWL833), a nonpeptide GLP-1R agonist. LY3502970 is a partial agonist, biased toward G protein activation over β-arrestin recruitment at the GLP-1R. The molecule is highly potent and selective against other class B G protein-coupled receptors (GPCRs) with a pharmacokinetic profile favorable for oral administration. A high-resolution structure of LY3502970 in complex with active-state GLP-1R revealed a unique binding pocket in the upper helical bundle where the compound is bound by the extracellular domain (ECD), extracellular loop 2, and transmembrane helices 1, 2, 3, and 7. This mechanism creates a distinct receptor conformation that may explain the partial agonism and biased signaling of the compound. Further, interaction between LY3502970 and the primate-specific Trp33 of the ECD informs species selective activity for the molecule. In efficacy studies, oral administration of LY3502970 resulted in glucose lowering in humanized GLP-1R transgenic mice and insulinotropic and hypophagic effects in nonhuman primates, demonstrating an effect size in both models comparable to injectable exenatide. Together, this work determined the molecular basis for the activity of an oral agent being developed for the treatment of type 2 diabetes mellitus, offering insights into the activation of class B GPCRs by nonpeptide ligands.
Organizational Affiliation:
Research Division, Chugai Pharmaceutical Co., Ltd., Gotemba, Shizuoka 412-8513, Japan.