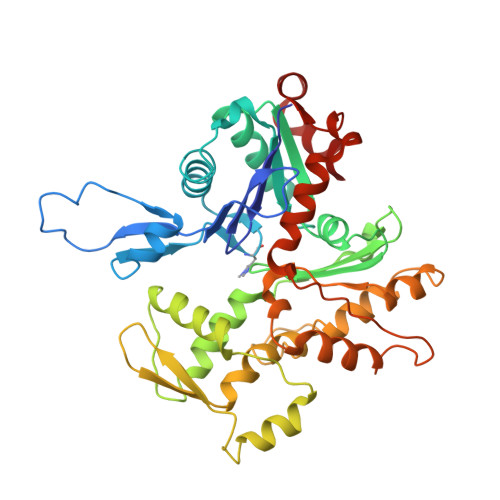
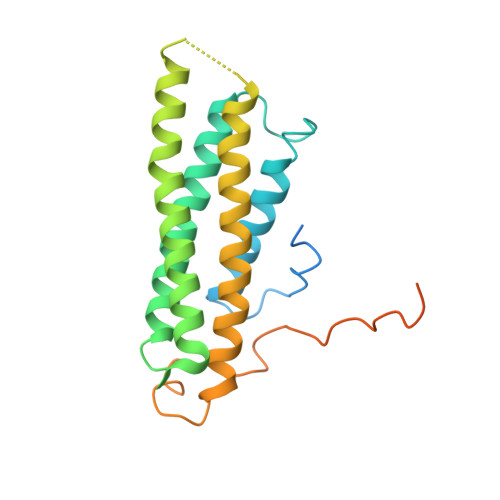
Structural basis of alpha E-catenin-F-actin catch bond behavior.
Xu, X.P., Pokutta, S., Torres, M., Swift, M.F., Hanein, D., Volkmann, N., Weis, W.I.(2020) Elife 9
- PubMed: 32915141
- DOI: https://doi.org/10.7554/eLife.60878
- Primary Citation of Related Structures:
6WVT - PubMed Abstract:
Cell-cell and cell-matrix junctions transmit mechanical forces during tissue morphogenesis and homeostasis. α-Catenin links cell-cell adhesion complexes to the actin cytoskeleton, and mechanical load strengthens its binding to F-actin in a direction-sensitive manner. Specifically, optical trap experiments revealed that force promotes a transition between weak and strong actin-bound states. Here, we describe the cryo-electron microscopy structure of the F-actin-bound αE-catenin actin-binding domain, which in solution forms a five-helix bundle. In the actin-bound structure, the first helix of the bundle dissociates and the remaining four helices and connecting loops rearrange to form the interface with actin. Deletion of the first helix produces strong actin binding in the absence of force, suggesting that the actin-bound structure corresponds to the strong state. Our analysis explains how mechanical force applied to αE-catenin or its homolog vinculin favors the strongly bound state, and the dependence of catch bond strength on the direction of applied force.
Organizational Affiliation:
Scintillon Institute, San Diego, United States.