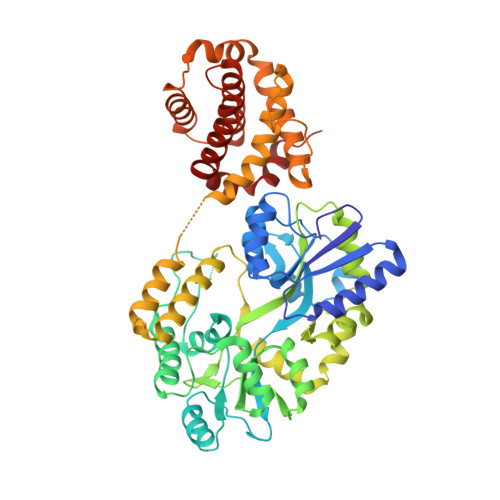
The structural basis of Bcl-2 mediated cell death regulation in hydra.
Banjara, S., D Sa, J., Hinds, M.G., Kvansakul, M.(2020) Biochem J 477: 3287-3297
- PubMed: 32776134
- DOI: https://doi.org/10.1042/BCJ20200556
- Primary Citation of Related Structures:
6WH0 - PubMed Abstract:
Apoptosis is regulated by evolutionarily conserved signaling pathways to remove damaged, diseased or unwanted cells. Proteins homologous to the B-cell lymphoma 2 (Bcl-2) family of proteins, the primary arbiters of mitochondrially mediated apoptosis, are encoded by the cnidarian Hydra vulgaris. We mapped interactions between pro-survival and pro-apoptotic Bcl-2 proteins of H. vulgaris by affinity measurements between Hy-Bcl-2-4, the sole confirmed pro-survival Bcl-2 protein, with BH3 motif peptides of two Bcl-2 proteins from hydra that displayed pro-apoptotic activity, Hy-Bak1 and Hy-BH3-only-2, and the BH3 motif peptide of the predicted pro-apoptotic protein Hy-Bax. In addition to peptides from hydra encoded pro-apoptotic proteins, Hy-Bcl-2-4 also engaged BH3 motif peptides from multiple human pro-apoptotic Bcl-2 proteins. Reciprocally, human pro-survival Bcl-2 proteins Bcl-2, Bcl-xL, Bcl-w, Mcl-1 and A1/Bfl-1 bound to BH3 spanning peptides from hydra encoded pro-apoptotic Hy-Bak1, Hy-BH3-only and Hy-Bax. The molecular details of the interactions were determined from crystal structures of Hy-Bcl-2-4 complexes with BH3 motif peptides of Hy-Bak1 and Hy-Bax. Our findings suggest that the Bcl-2 family in hydra may function in a manner analogous to the Bcl-2 family in humans, and less like the worm Caenorhabditis elegans where evolutionary gene deletion has simplified the apoptotic program. Combined, our results demonstrate the powerful conservation of the interaction pattern between hydra and human Bcl-2 family members. Furthermore, our data reveal mechanistic differences in the mode of binding between hydra and sponges such as Geodia cydonium, with hydra encoded Bcl-2 resembling the more promiscuous pro-apoptotic Bcl-2 members found in mammals compared with its sponge counterpart.
Organizational Affiliation:
Department of Biochemistry and Genetics, La Trobe Institute for Molecular Science, La Trobe University, Melbourne, Victoria 3086, Australia.