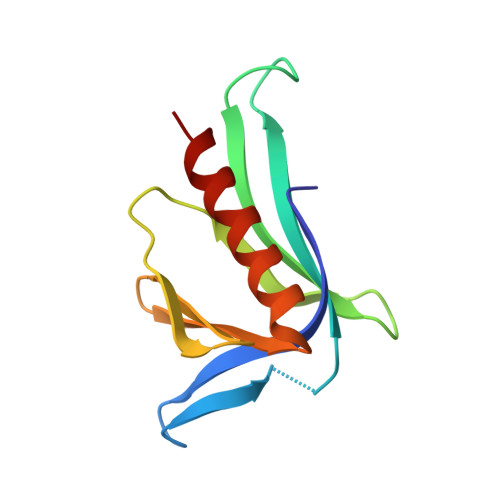
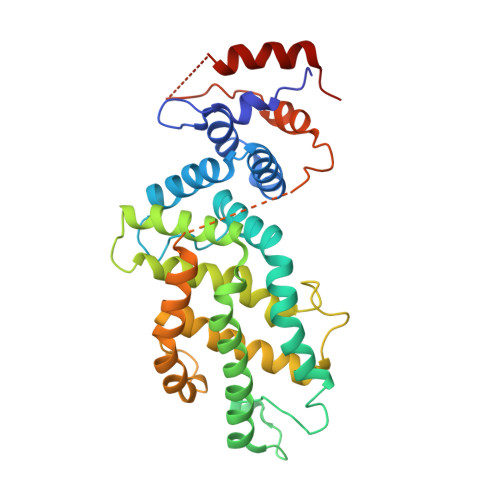
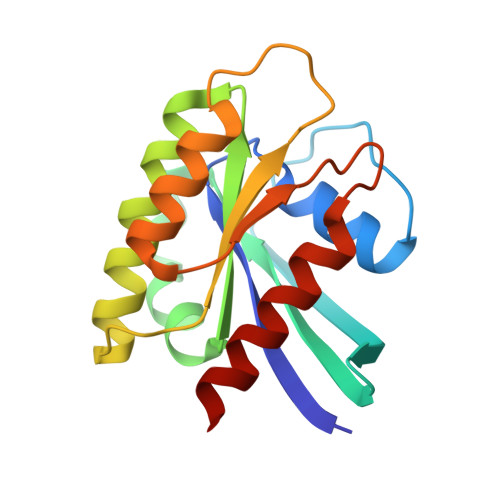
Structural Insights into the SPRED1-Neurofibromin-KRAS Complex and Disruption of SPRED1-Neurofibromin Interaction by Oncogenic EGFR.
Yan, W., Markegard, E., Dharmaiah, S., Urisman, A., Drew, M., Esposito, D., Scheffzek, K., Nissley, D.V., McCormick, F., Simanshu, D.K.(2020) Cell Rep 32: 107909-107909
- PubMed: 32697994
- DOI: https://doi.org/10.1016/j.celrep.2020.107909
- Primary Citation of Related Structures:
6V65, 6V6F - PubMed Abstract:
Sprouty-related, EVH1 domain-containing (SPRED) proteins negatively regulate RAS/mitogen-activated protein kinase (MAPK) signaling following growth factor stimulation. This inhibition of RAS is thought to occur primarily through SPRED1 binding and recruitment of neurofibromin, a RasGAP, to the plasma membrane. Here, we report the structure of neurofibromin (GTPase-activating protein [GAP]-related domain) complexed with SPRED1 (EVH1 domain) and KRAS. The structure provides insight into how the membrane targeting of neurofibromin by SPRED1 allows simultaneous interaction with activated KRAS. SPRED1 and NF1 loss-of-function mutations occur across multiple cancer types and developmental diseases. Analysis of the neurofibromin-SPRED1 interface provides a rationale for mutations observed in Legius syndrome and suggests why SPRED1 can bind to neurofibromin but no other RasGAPs. We show that oncogenic EGFR(L858R) signaling leads to the phosphorylation of SPRED1 on serine 105, disrupting the SPRED1-neurofibromin complex. The structural, biochemical, and biological results provide new mechanistic insights about how SPRED1 interacts with neurofibromin and regulates active KRAS levels in normal and pathologic conditions.
Organizational Affiliation:
NCI RAS Initiative, Cancer Research Technology Program, Frederick National Laboratory for Cancer Research, Leidos Biomedical Research, Inc., Frederick, MD 21701, USA.