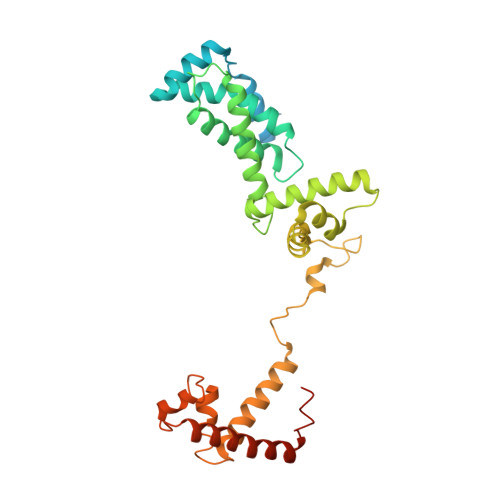
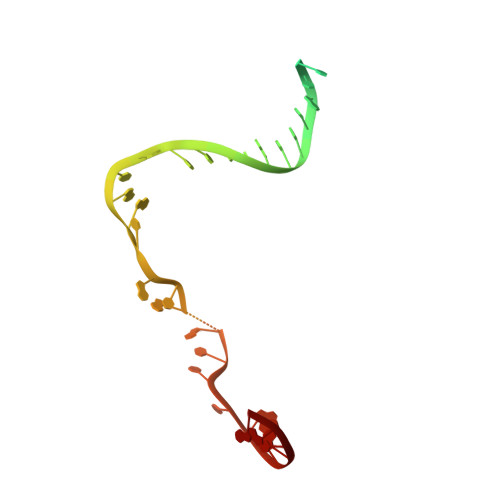
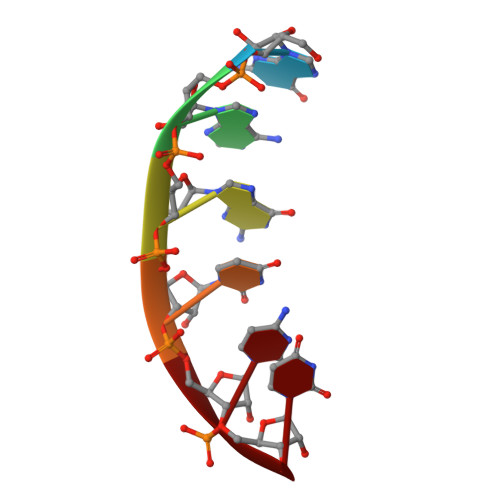
Structural Insights into Transcription Initiation from De Novo RNA Synthesis to Transitioning into Elongation.
Zuo, Y., De, S., Feng, Y., Steitz, T.A.(2020) iScience 23: 101445-101445
- PubMed: 32829286
- DOI: https://doi.org/10.1016/j.isci.2020.101445
- Primary Citation of Related Structures:
6UTV, 6UTW, 6UTX, 6UTY, 6UTZ, 6UU0, 6UU1, 6UU2, 6UU3, 6UU4, 6UU5, 6UU6, 6UU7, 6UU8, 6UU9, 6UUA, 6UUB, 6UUC - PubMed Abstract:
In bacteria, the dissociable σ subunit of the RNA polymerase (RNAP) is responsible for initiating RNA synthesis from specific DNA sites. As nascent RNA grows, downstream DNA unwinds and is pulled into the RNAP, causing stress accumulation and initiation complex destabilization. Processive transcription elongation requires at least partial separation of the σ factor from the RNAP core enzyme. Here, we present a series of transcription complexes captured between the early initiation and elongation phases via in-crystal RNA synthesis and cleavage. Crystal structures of these complexes indicate that stress accumulation during transcription initiation is not due to clashing of the growing nascent RNA with the σ 3.2 loop, but results from scrunching of the template strand DNA that is contained inside the RNAP by the σ 3 domain. Our results shed light on how scrunching of template-strand DNA drives both abortive initiation and σ-RNAP core separation to transition transcription from initiation to elongation.
Organizational Affiliation:
Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT 06520, USA.