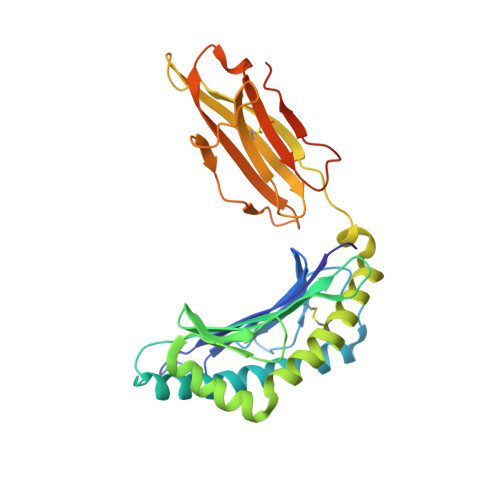
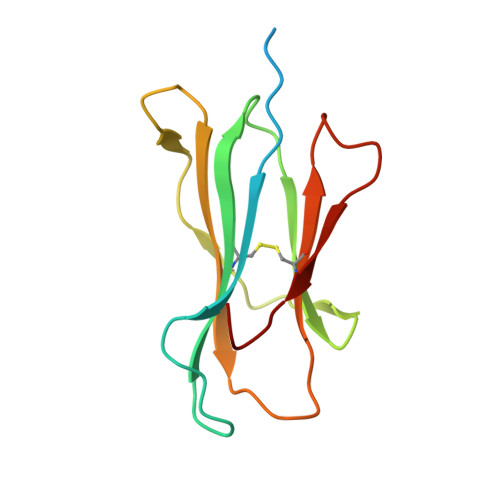
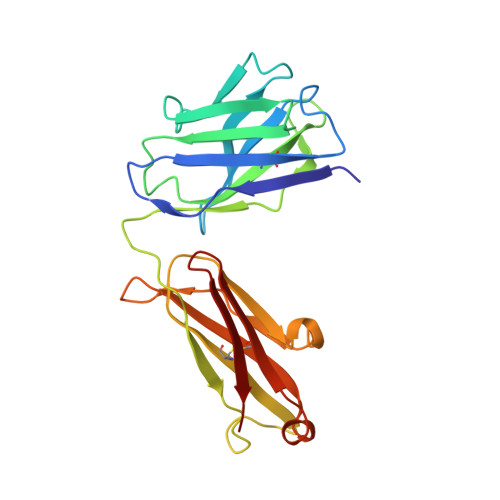
Structural engineering of chimeric antigen receptors targeting HLA-restricted neoantigens.
Hwang, M.S., Miller, M.S., Thirawatananond, P., Douglass, J., Wright, K.M., Hsiue, E.H., Mog, B.J., Aytenfisu, T.Y., Murphy, M.B., Aitana Azurmendi, P., Skora, A.D., Pearlman, A.H., Paul, S., DiNapoli, S.R., Konig, M.F., Bettegowda, C., Pardoll, D.M., Papadopoulos, N., Kinzler, K.W., Vogelstein, B., Zhou, S., Gabelli, S.B.(2021) Nat Commun 12: 5271-5271
- PubMed: 34489470
- DOI: https://doi.org/10.1038/s41467-021-25605-4
- Primary Citation of Related Structures:
6UJ7, 6UJ8, 6UJ9, 7KGU - PubMed Abstract:
Chimeric antigen receptor (CAR) T cells have emerged as a promising class of therapeutic agents, generating remarkable responses in the clinic for a subset of human cancers. One major challenge precluding the wider implementation of CAR therapy is the paucity of tumor-specific antigens. Here, we describe the development of a CAR targeting the tumor-specific isocitrate dehydrogenase 2 (IDH2) with R140Q mutation presented on the cell surface in complex with a common human leukocyte antigen allele, HLA-B*07:02. Engineering of the hinge domain of the CAR, as well as crystal structure-guided optimization of the IDH2 R140Q -HLA-B*07:02-targeting moiety, enhances the sensitivity and specificity of CARs to enable targeting of this HLA-restricted neoantigen. This approach thus holds promise for the development and optimization of immunotherapies specific to other cancer driver mutations that are difficult to target by conventional means.
Organizational Affiliation:
Ludwig Center, Sidney Kimmel Comprehensive Cancer Center, Johns Hopkins University School of Medicine, Baltimore, MD, USA.