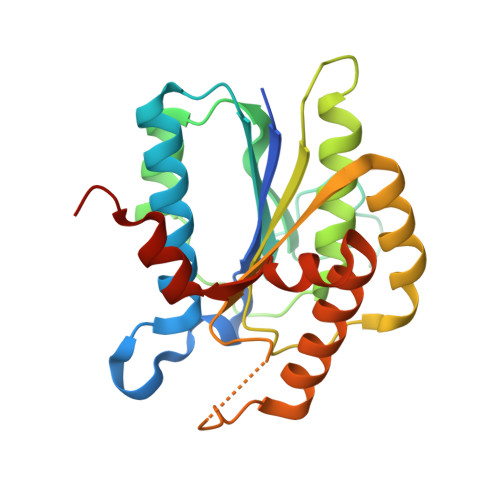
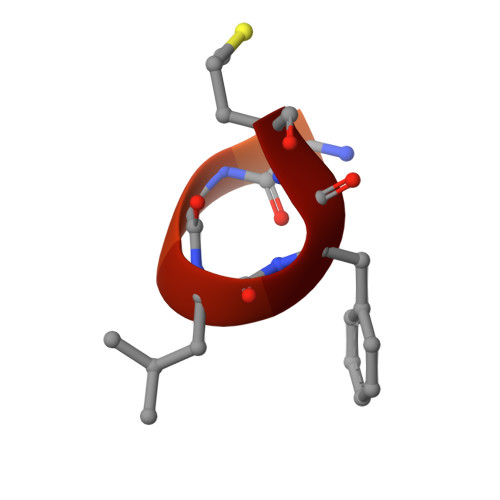
Ligand binding characteristics of the Ku80 von Willebrand domain.
Kim, K., Min, J., Kirby, T.W., Gabel, S.A., Pedersen, L.C., London, R.E.(2019) DNA Repair (Amst) 85: 102739-102739
- PubMed: 31733588
- DOI: https://doi.org/10.1016/j.dnarep.2019.102739
- Primary Citation of Related Structures:
6TYT, 6TYU, 6TYV, 6TYW, 6TYX, 6TYZ - PubMed Abstract:
The N-terminal von Willebrand domain of Ku80 supports interactions with a Ku binding motif (KBM) that has been identified in at least three other DNA repair proteins: the non-homologous end joining (NHEJ) scaffold APLF, the modulator of retrovirus infection, MRI, and the Werner syndrome protein (WRN). A second, more recently identified Ku binding motif present in XLF and several other proteins (KBMX) has also been reported to interact with this domain. The isolated Ku80 von Willebrand antigen domain (vWA) from Xenopus laevis has a sequence that is 60% identical with the human domain, is readily expressed and has been used to investigate these interactions. Structural characterization of the complexes formed with the KBM motifs in human APLF, MRI, and WRN identify a conserved binding site that is consistent with previously-reported mutational studies. In contrast with the KBM binding site, structural studies indicate that the KBMX site is occluded by a distorted helix. Fluorescence polarization and 19 F NMR studies of a fluorinated XLF C-terminal peptide failed to indicate any interaction with the frog vWA. It was hypothesized that availability of this binding site is conditional, i.e., dependent on specific experimental conditions or other repair factors to make the site available for binding. Modulating the fraction of KBMX-accessible binding site mutationally demonstrated that the more open site is capable of binding the KBMX XLF motif peptide. It is suggested that the conditional nature of KBMX binding limits formation of non-productive complexes so that activation-dependent site availability can more optimally support advancing the synapsis process.
Organizational Affiliation:
Genome Integrity and Structural Biology Laboratory, National Institute of Environment and Health Sciences, National Institutes of Health, Research Triangle Park, NC, 27709, USA.