Dual Specificity PDZ- and 14-3-3-Binding Motifs: A Structural and Interactomics Study.
Gogl, G., Jane, P., Caillet-Saguy, C., Kostmann, C., Bich, G., Cousido-Siah, A., Nyitray, L., Vincentelli, R., Wolff, N., Nomine, Y., Sluchanko, N.N., Trave, G.(2020) Structure 28: 747-759.e3
- PubMed: 32294469
- DOI: https://doi.org/10.1016/j.str.2020.03.010
- Primary Citation of Related Structures:
6TWQ, 6TWU, 6TWX, 6TWY, 6TWZ - PubMed Abstract:
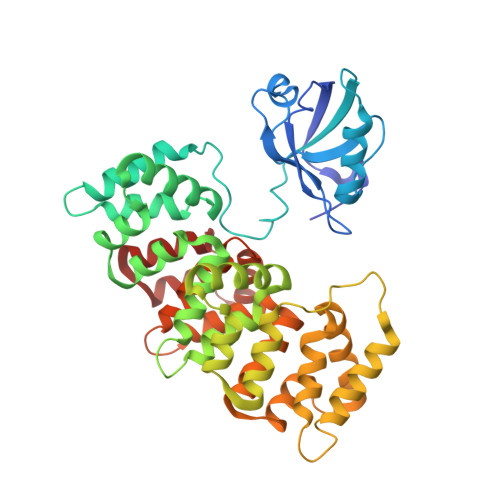
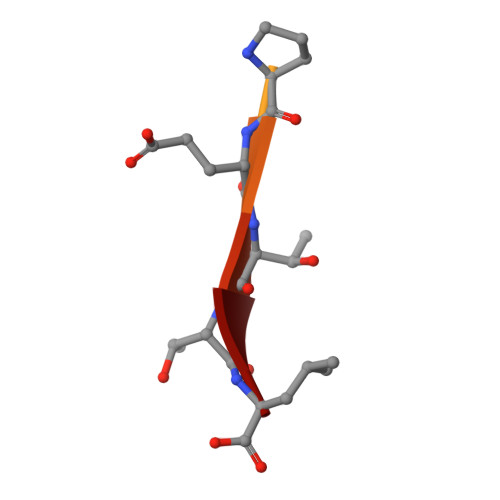
Protein-protein interaction motifs are often alterable by post-translational modifications. For example, 19% of predicted human PDZ domain-binding motifs (PBMs) have been experimentally proven to be phosphorylated, and up to 82% are theoretically phosphorylatable. Phosphorylation of PBMs may drastically rewire their interactomes, by altering their affinities for PDZ domains and 14-3-3 proteins. The effect of phosphorylation is often analyzed by performing "phosphomimetic" mutations. Here, we focused on the PBMs of HPV16-E6 viral oncoprotein and human RSK1 kinase. We measured the binding affinities of native, phosphorylated, and phosphomimetic variants of both PBMs toward the 266 human PDZ domains. We co-crystallized all the motif variants with a selected PDZ domain to characterize the structural consequence of the different modifications. Finally, we elucidated the structural basis of PBM capture by 14-3-3 proteins. This study provides novel atomic and interactomic insights into phosphorylatable dual specificity motifs and the differential effects of phosphorylation and phosphomimetic approaches.
Organizational Affiliation:
Equipe Labellisee Ligue 2015, Department of Integrated Structural Biology, Institut de Genetique et de Biologie Moleculaire et Cellulaire (IGBMC), INSERM U1258/CNRS UMR 7104/Universite de Strasbourg, 1 rue Laurent Fries, BP 10142, 67404 Illkirch, France. Electronic address: goglg@igbmc.fr.