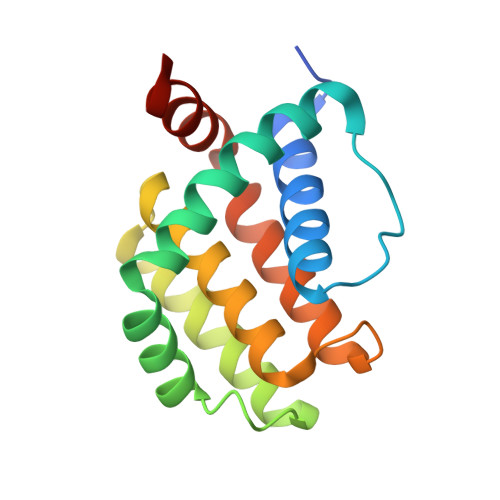
Structural insight into tanapoxvirus-mediated inhibition of apoptosis.
Suraweera, C.D., Anasir, M.I., Chugh, S., Javorsky, A., Impey, R.E., Hasan Zadeh, M., Soares da Costa, T.P., Hinds, M.G., Kvansakul, M.(2020) FEBS J 287: 3733-3750
- PubMed: 32412687
- DOI: https://doi.org/10.1111/febs.15365
- Primary Citation of Related Structures:
6TQP, 6TQQ, 6TRR - PubMed Abstract:
Premature programmed cell death or apoptosis of cells is a strategy utilized by multicellular organisms to counter microbial threats. Tanapoxvirus (TANV) is a large double-stranded DNA virus belonging to the poxviridae that causes mild monkeypox-like infections in humans and primates. TANV encodes for a putative apoptosis inhibitory protein 16L. We show that TANV16L is able to bind to a range of peptides spanning the BH3 motif of human proapoptotic Bcl-2 proteins and is able to counter growth arrest of yeast induced by human Bak and Bax. We then determined the crystal structures of TANV16L bound to three identified interactors, Bax, Bim and Puma BH3. TANV16L adopts a globular Bcl-2 fold comprising 7 α-helices and utilizes the canonical Bcl-2 binding groove to engage proapoptotic host cell Bcl-2 proteins. Unexpectedly, TANV16L is able to adopt both a monomeric and a domain-swapped dimeric topology where the α1 helix from one protomer is swapped into a neighbouring unit. Despite adopting two different oligomeric forms, the canonical ligand binding groove in TANV16L remains unchanged from monomer to domain-swapped dimer. Our results provide a structural and mechanistic basis for tanapoxvirus-mediated inhibition of host cell apoptosis and reveal the capacity of Bcl-2 proteins to adopt differential oligomeric states whilst maintaining the canonical ligand binding groove in an unchanged state. DATABASE: Structural data are available in the Protein Data Bank (PDB) under the accession numbers 6TPQ, 6TQQ and 6TRR.
Organizational Affiliation:
Department of Biochemistry and Genetics, La Trobe Institute for Molecular Science, La Trobe University, Melbourne, Australia.