Demonstrating Ligandability of the LC3A and LC3B Adapter Interface.
Hartmann, M., Huber, J., Kramer, J.S., Heering, J., Pietsch, L., Stark, H., Odadzic, D., Bischoff, I., Furst, R., Schroder, M., Akutsu, M., Chaikuad, A., Dotsch, V., Knapp, S., Biondi, R.M., Rogov, V.V., Proschak, E.(2021) J Med Chem 64: 3720-3746
- PubMed: 33769048
- DOI: https://doi.org/10.1021/acs.jmedchem.0c01564
- Primary Citation of Related Structures:
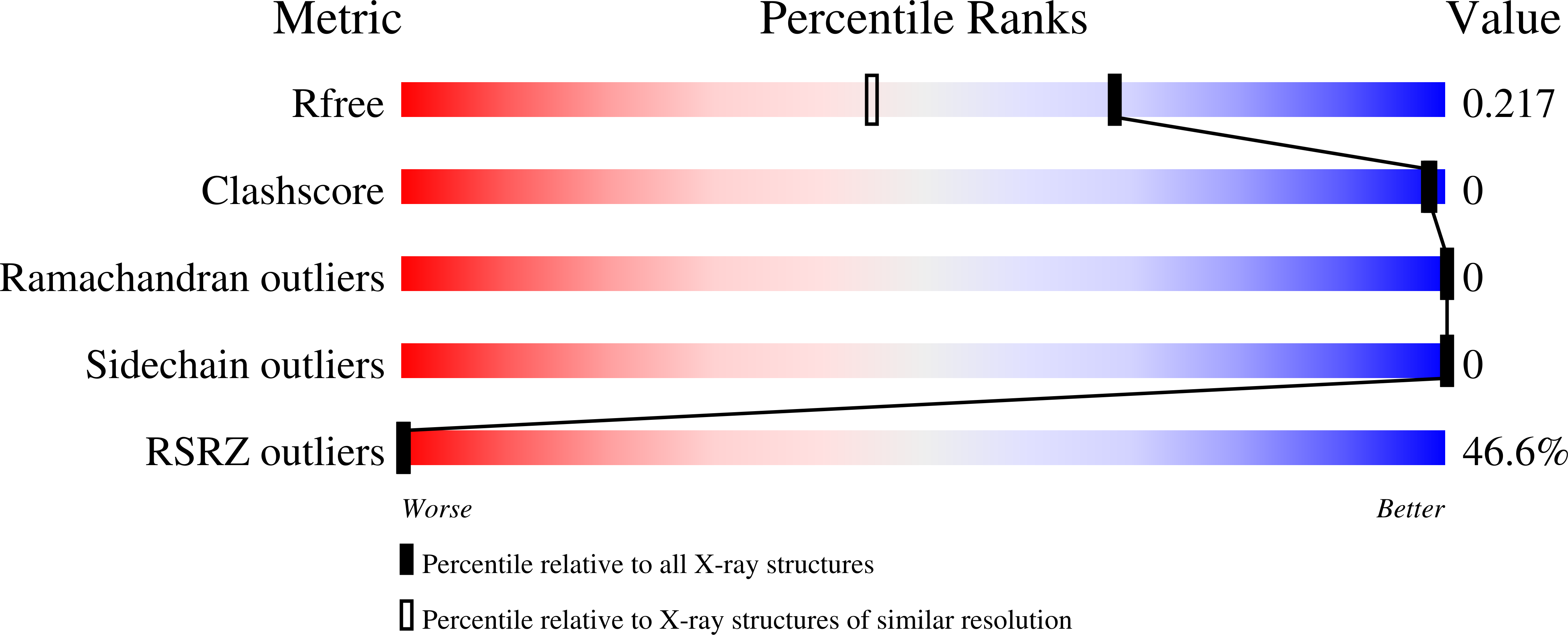
6TBE - PubMed Abstract:
Autophagy is the common name for a number of lysosome-based degradation pathways of cytosolic cargos. The key components of autophagy are members of Atg8 family proteins involved in almost all steps of the process, from autophagosome formation to their selective fusion with lysosomes. In this study, we show that the homologous members of the human Atg8 family proteins, LC3A and LC3B, are druggable by a small molecule inhibitor novobiocin. Structure-activity relationship (SAR) studies of the 4-hydroxy coumarin core scaffold were performed, supported by a crystal structure of the LC3A dihydronovobiocin complex. The study reports the first nonpeptide inhibitors for these protein interaction targets and will lay the foundation for the development of more potent chemical probes for the Atg8 protein family which may also find applications for the development of autophagy-mediated degraders (AUTACs).
Organizational Affiliation:
Institute of Pharmaceutical Chemistry, Goethe-University Frankfurt, Max-von-Laue-Str. 9, 60438 Frankfurt, Germany.