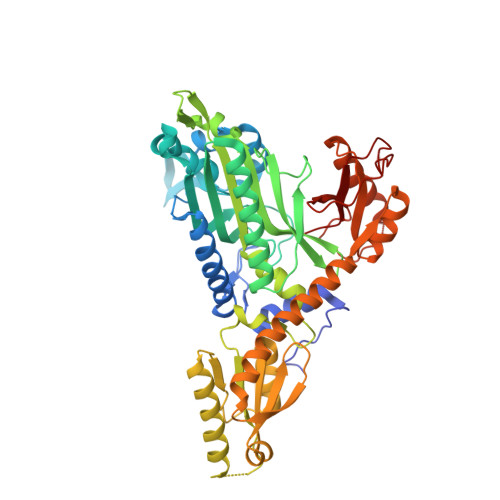
Crystal Structure of Prolyl-tRNA synthetase (ProRS, Proline--tRNA ligase) from Plasmodium falciparum in complex with NCP-26 and L-Proline
Johansson, C., Wang, J., Tye, M., Payne, N.C., Mazitschek, R., Thompson, A., Arrowsmith, C.H., Bountra, C., Edwards, A., Oppermann, U.C.T.To be published.