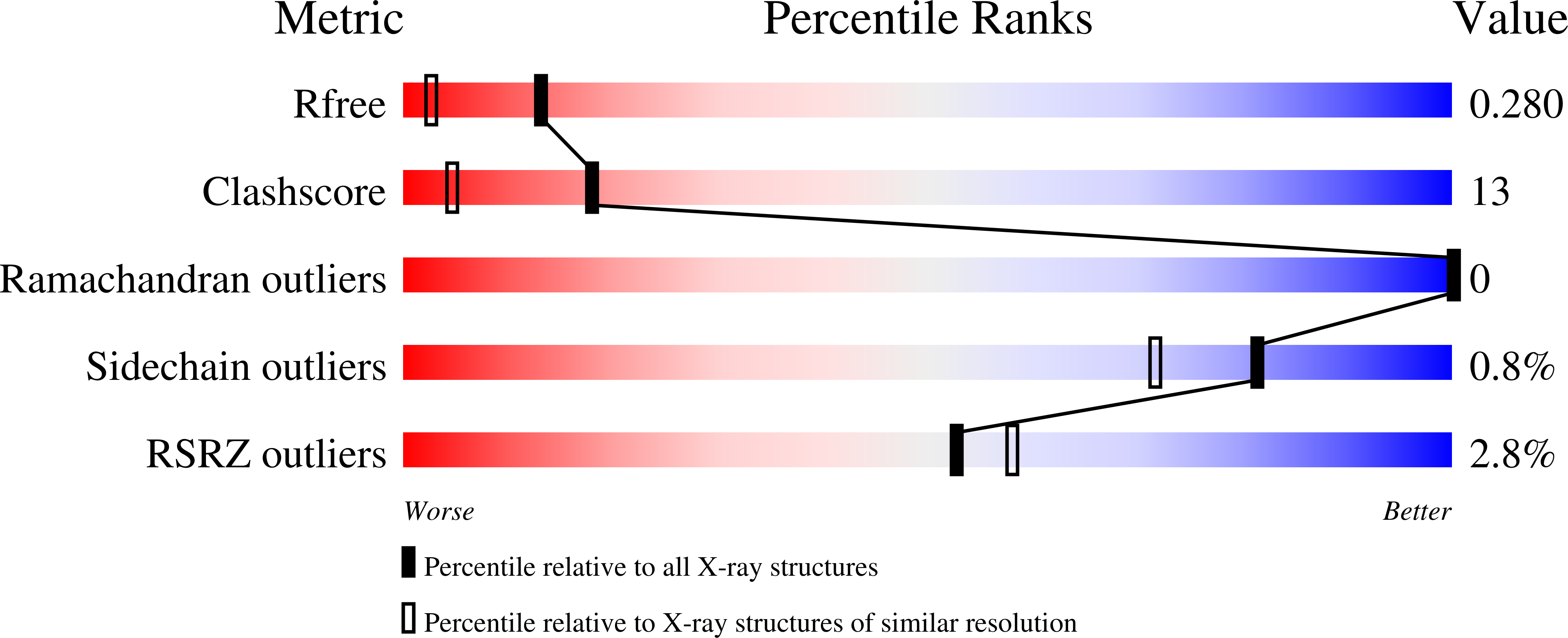
Structural analysis of the reducing-end xylose-releasing exo-oligoxylanase Rex8A from Paenibacillus barcinonensis BP-23 deciphers its molecular specificity.
Jimenez-Ortega, E., Valenzuela, S., Ramirez-Escudero, M., Pastor, F.J., Sanz-Aparicio, J.(2020) FEBS J 287: 5362-5374
- PubMed: 32352213
- DOI: https://doi.org/10.1111/febs.15332
- Primary Citation of Related Structures:
6SHY, 6SRD, 6SUD, 6TO0, 6TOW, 6TPP, 6TRH - PubMed Abstract:
Reducing-end xylose-releasing exo-oligoxylanases (Rex) are GH8 enzymes that depolymerize xylooligosaccharides complementing xylan degradation by endoxylanases in an exo manner. We have studied Paenibacillus barcinonensis Rex8A and showed the release of xylose from xylooligomers decorated with methylglucuronic acid (UXOS) or with arabinose (AXOS). This gives the enzyme a distinctive trait among known Rex, which show activity only on linear xylooligosaccharides. The structure of the enzyme has been solved by X-ray crystallography showing a (α/α) 6 folding common to GH8 enzymes. Analysis of inactived Rex8A-E70A complexed with xylotetraose revealed the existence of at least four binding subsites in Rex8A, with the oligosaccharide occupying subsites -3 to +1. The enzyme shows an extended Leu320-His321-Pro322 loop, common to other Rex, which blocks the binding of longer substrates to positive subsites further than +1 and seems responsible for the lack or diminished activity of Rex enzymes on xylan. Mutants with smaller residues in this loop failed to increase Rex8A activity on the polymer. Analysis of the complexes with AXOS showed the accommodation of arabinose at subsite -2, which cannot be allocated at subsite -1. Arabinose substitutions at the xylose O2 or O3 are accommodated by hydrophobic interaction and seem tolerated rather than recognized by Rex8A. A strained binding of the branch is facilitated by the lack of direct polar interactions of the xylose occupying this subsite, its water-mediated links allowing some conformational flexibility of the sugar. The plasticity of Rex8A is a notable property of the enzyme for its application in xylan deconstruction and upgrading. DATABASE: Structural data are available in PDB database under the accession numbers 6SRD (native form), 6TPP (E70A mutant in complex with EDO), 6TOW (E70A in complex with Xyl4), 6SUD (L320A mutant in complex with xylose), 6SHY (L320A/H321S double mutant in complex with EDO), 6TO0 (E70A in complex with AX3), and 6TRH (E70A in complex with AX4).
Organizational Affiliation:
Macromolecular Crystallography and Structural Biology Department, Institute of Physical-Chemistry 'Rocasolano', CSIC, Madrid, Spain.