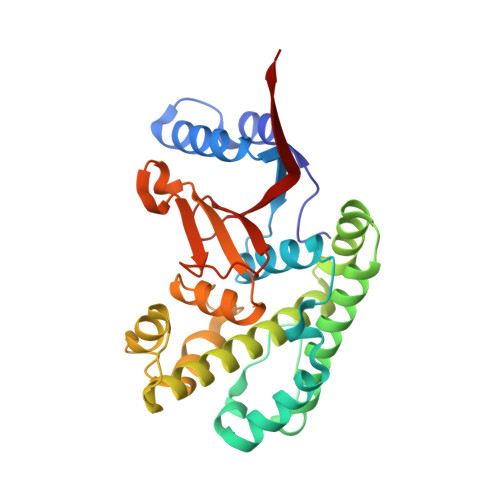
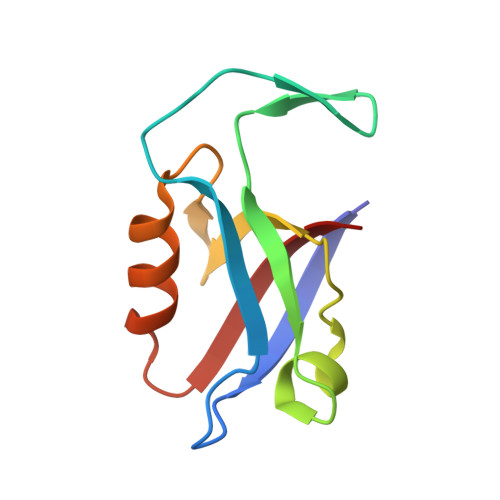
Regulation of the endosomal SNX27-retromer by OTULIN.
Stangl, A., Elliott, P.R., Pinto-Fernandez, A., Bonham, S., Harrison, L., Schaub, A., Kutzner, K., Keusekotten, K., Pfluger, P.T., El Oualid, F., Kessler, B.M., Komander, D., Krappmann, D.(2019) Nat Commun 10: 4320-4320
- PubMed: 31541095
- DOI: https://doi.org/10.1038/s41467-019-12309-z
- Primary Citation of Related Structures:
6SAK - PubMed Abstract:
OTULIN (OTU Deubiquitinase With Linear Linkage Specificity) specifically hydrolyzes methionine1 (Met1)-linked ubiquitin chains conjugated by LUBAC (linear ubiquitin chain assembly complex). Here we report on the mass spectrometric identification of the OTULIN interactor SNX27 (sorting nexin 27), an adaptor of the endosomal retromer complex responsible for protein recycling to the cell surface. The C-terminal PDZ-binding motif (PDZbm) in OTULIN associates with the cargo-binding site in the PDZ domain of SNX27. By solving the structure of the OTU domain in complex with the PDZ domain, we demonstrate that a second interface contributes to the selective, high affinity interaction of OTULIN and SNX27. SNX27 does not affect OTULIN catalytic activity, OTULIN-LUBAC binding or Met1-linked ubiquitin chain homeostasis. However, via association, OTULIN antagonizes SNX27-dependent cargo loading, binding of SNX27 to the VPS26A-retromer subunit and endosome-to-plasma membrane trafficking. Thus, we define an additional, non-catalytic function of OTULIN in the regulation of SNX27-retromer assembly and recycling to the cell surface.
Organizational Affiliation:
Research Unit Cellular Signal Integration, Institute of Molecular Toxicology and Pharmacology, Helmholtz Zentrum München, Ingolstaedter Landstrasse 1, 85764, Neuherberg, Germany.