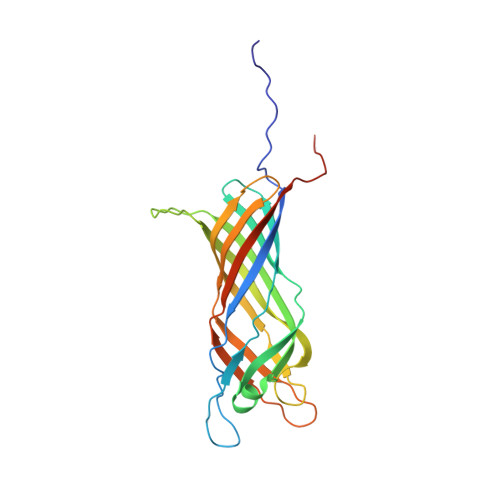
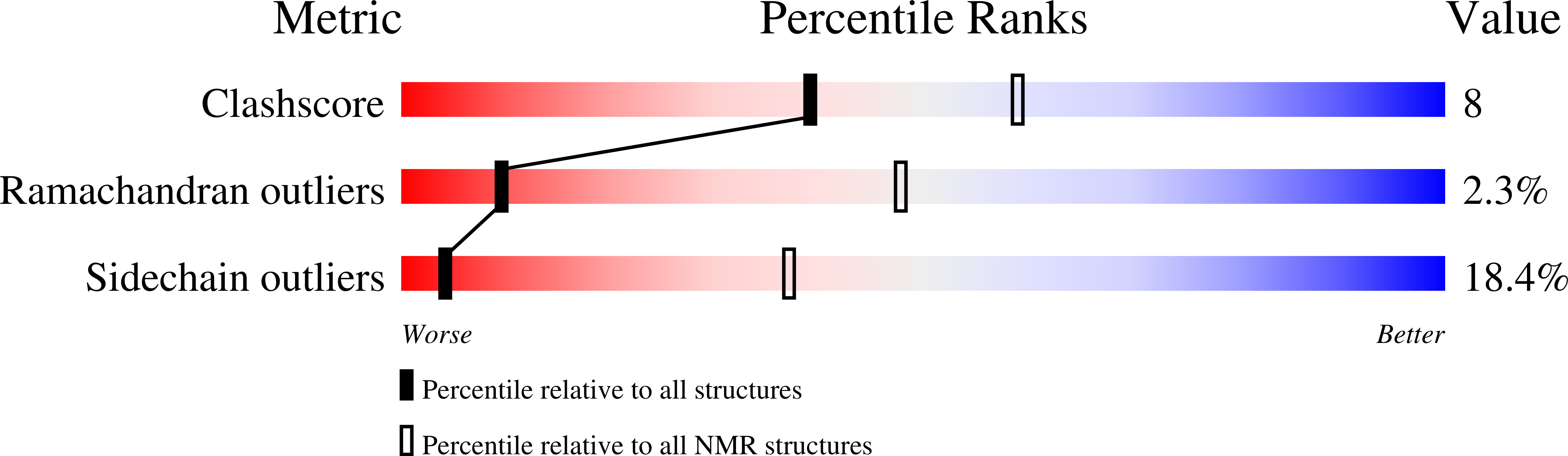A beta-barrel for oil transport through lipid membranes: Dynamic NMR structures of AlkL.
Schubeis, T., Le Marchand, T., Daday, C., Kopec, W., Tekwani Movellan, K., Stanek, J., Schwarzer, T.S., Castiglione, K., de Groot, B.L., Pintacuda, G., Andreas, L.B.(2020) Proc Natl Acad Sci U S A 117: 21014-21021
- PubMed: 32817429
- DOI: https://doi.org/10.1073/pnas.2002598117
- Primary Citation of Related Structures:
6QAM, 6QWR - PubMed Abstract:
The protein AlkL is known to increase permeability of the outer membrane of bacteria for hydrophobic molecules, yet the mechanism of transport has not been determined. Differing crystal and NMR structures of homologous proteins resulted in a controversy regarding the degree of structure and the role of long extracellular loops. Here we solve this controversy by determining the de novo NMR structure in near-native lipid bilayers, and by accessing structural dynamics relevant to hydrophobic substrate permeation through molecular-dynamics simulations and by characteristic NMR relaxation parameters. Dynamic lateral exit sites large enough to accommodate substrates such as carvone or octane occur through restructuring of a barrel extension formed by the extracellular loops.
Organizational Affiliation:
Centre de Résonance Magnétique Nucléaire à Très Hauts Champs de Lyon (FRE 2034-CNRS, Université Claude Bernard Lyon 1, École Normale Supérieure Lyon), Université de Lyon, 69100 Villeurbanne, France.